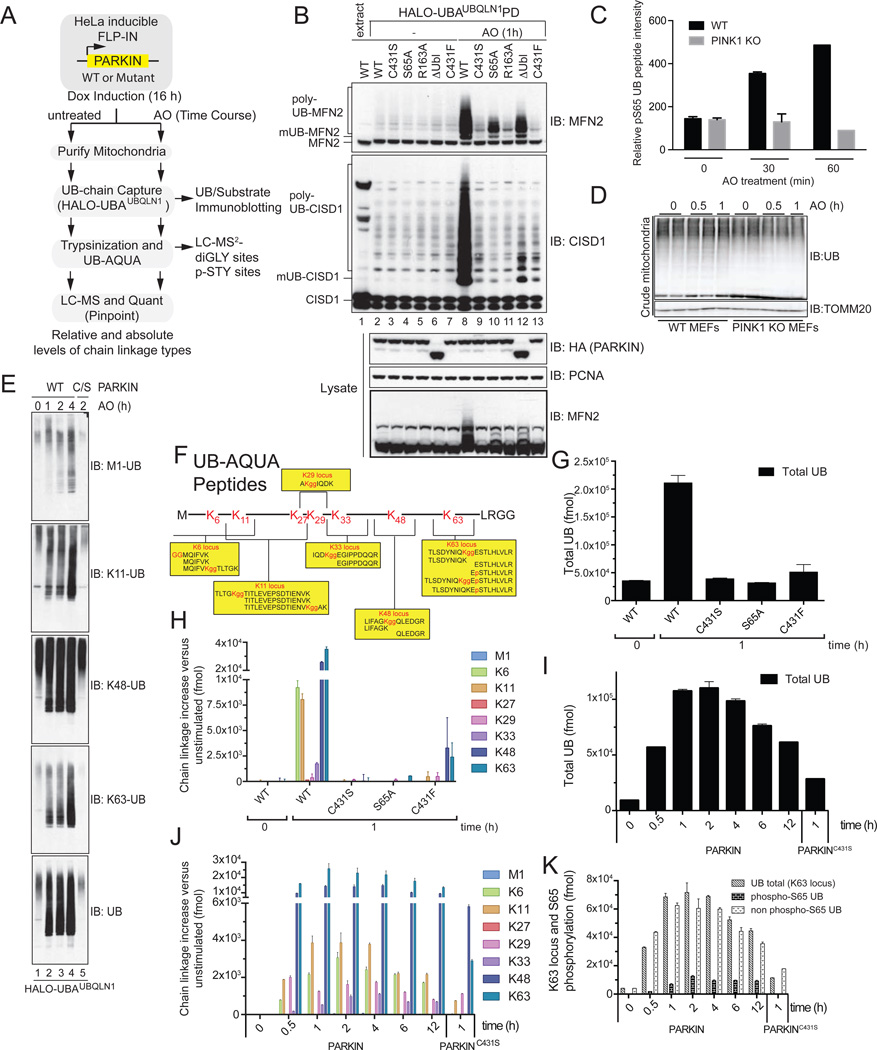
Figure 1. Analysis of PARKIN and PINK1-dependent UB chain linkage formation on mitochondria in response to depolarization.
(A) Schematic UB-AQUA proteomics workflow.
(B) Ubiquitylated proteins present on purified mitochondria from HeLa Flp-In T-REx cells after depolarization were captured using immobilized Halo-UBAUBQLN1 prior to immunoblotting with α-MFN2 and α-CISD1.
(C,D) PINK1-dependent phosphorylation of S65 in UB. (C) Mitochondria from WT or PINK1−/− MEFs were subjected to 10-plex TMT at the indicated times post depolarization with AO and the S65 UB tryptic peptide quantified. (D) Total UB levels on mitochondria from Panel C.
(E) Mitochondria from cells subjected to the indicated treatments was used for poly-UB capture using Halo-UBAUBQLN1 and proteins immunoblotted with linkage specific antibodies.
(F) AQUA peptides used to quantify diGLY and phosphopeptides [after (Phu et al., 2011)].
(G–J) UB-AQUA proteomics of total UB (G) and individual UB chain linkage types (H) associated with mitochondria in response to depolarization with AO for 1 h in the presence of the indicated PARKIN protein (G,H) and over time (I,J). Error bars represent triplicate measurements (+/− SEM).
(K) UB-AQUA for total UB (K63 locus) and p-S65 UB accumulation on mitochondria in response to depolarization in the presence of WT or C431S PARKIN, and error bars represent triplicate measurements (+/− SEM).