Figure 3.
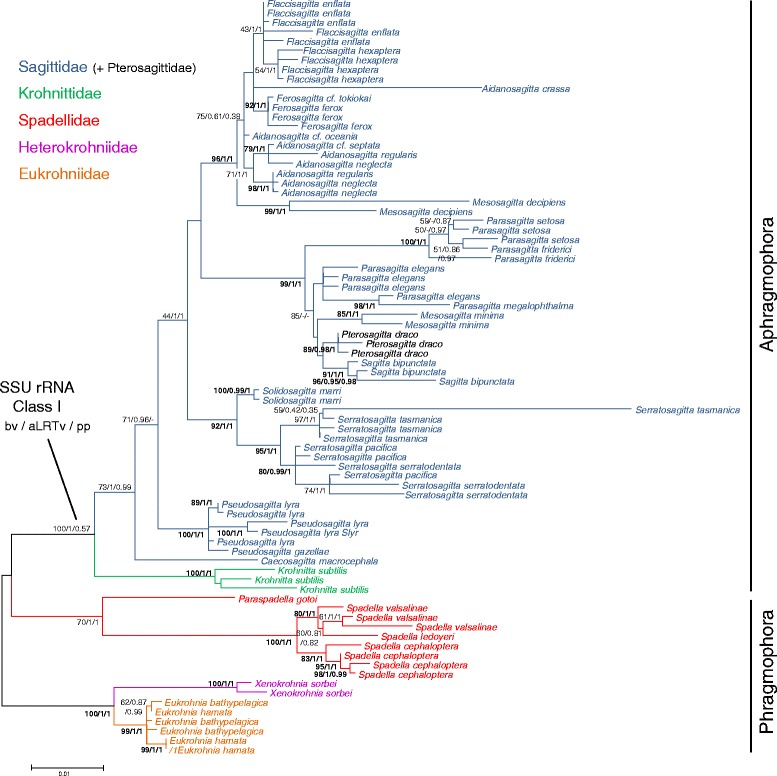
Maximum likelihood tree calculated from SSU rRNA Class I by the Mega program with the K2P + Γ model. The alignment of 80 sequences of LSU rRNA was 1679 base pairs long. The lnL value of this optimal tree was −7733.81. Support values obtained using different reconstruction approaches are indicated at nodes in the following order: maximum Likelihood bootstrap probabilities (bv), approximative likelihood ratio test (aLRTv) and Bayesian posterior probabilities (pp). Support values are displayed when bv/aLRTv ≥75 or pp ≥0.85. Node absence in a given method is indicated by -. Tree was rooted on the monophyletic assemblage consisting of Eukrohniidae + Heterokrohniidae. This rooting was also observed in most analyses when using the midpoint rooting function.
