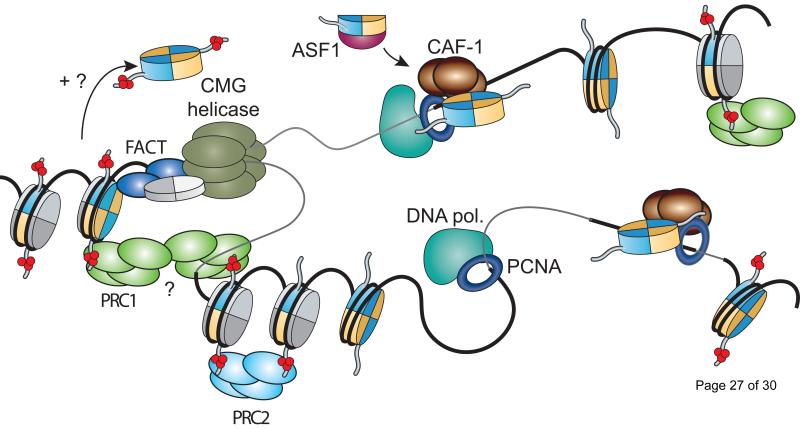
Figure 1.
Histone dynamics and inheritance of epigenetic information at the replication fork, as exemplified by the methylation of histone H3 on lysine 27. De novo nucleosome assembly proceeds through the nuclear import of histone H3-H4 dimers via the ASF1 histone chaperone. Differential thermodynamic affinities towards histones facilitate the transfer of these predominantly unmodified histones to the PCNA-bound CAF-1. The latter facilitates the formation and deposition of stable (H3.1-H4)2 tetramers to be completed by the addition of two juxtaposed H2A-H2B dimers. Nucleosomes encountering the replication fork are transiently bound and dissociated by the MCM2 subunit of the CMG replicative helicase as well as by the histone chaperone FACT. The exact mechanism by which these multi-PTM decorated histone tetramers segregate onto nascent DNA strands remains to be fully elucidated. Polycomb proteins persist at the replication fork through internucleosomal contacts. Only histone H3 N-terminal tails marked on lysine 27 are shown for simplicity. Histones H3, H4, and H2A-H2B are shown in blue, yellow and grey, respectively. Methyl marks are shown in red. Figure is adapted from [2].