Abstract
The Golgi apparatus is a multi-compartment central sorting station at the intersection of secretory and endocytic vesicular traffic. The mechanisms that permit cargo-loaded transport vesicles from different origins to selectively access different Golgi compartments are incompletely understood. Here, we develop a re-routing and capture assay to investigate systematically the vesicle-tethering activities of ten widely conserved golgin coiled-coil proteins. We find that subsets of golgins with distinct localizations on the Golgi surface have capture activities toward vesicles of different origins. These findings demonstrate that golgins act as tethers in vivo, and hence the specificity we find to be encoded in this tethering is likely to make a major contribution to the organization of membrane traffic at the Golgi apparatus.
The functionality of the membrane-bound organelles of eukaryotic cells is determined by their constituent proteins and lipids. A major determinant of organelle composition in the endomembrane system is the highly selective transfer of cargo-laden vesicles between compartments. Not only should the correct cargo be collected at the origin, but the transport vesicle must be selectively delivered to the correct destination. SNARE proteins on the vesicle and destination organelle assemble to drive membrane fusion (1). The diversity of cellular SNAREs, their differential localization, and their selective pairwise interactions implicate them in contributing specificity to membrane traffic (2-4). However, the relatively small size of SNAREs means they can only interact after a vesicle is closely apposed to its potential destination. A process called ‘tethering’ is thought to attach the vesicle to the organelle before SNARE complex assembly (5-7). The degree to which organelle tethers confer specificity rather than efficiency to membrane traffic is presently unclear.
To investigate this problem, we focused on the Golgi apparatus, the central sorting station in the endomembrane system (8). The Golgi is a stack of distinct cisternae arranged from cis to trans, and particular vesicles appear to preferentially fuse with different cisternae within the stack. For example, vesicles derived from the endoplasmic reticulum (ER) deliver cargo to the cis face of the Golgi, while those from the endocytic pathway deliver cargo to the trans Golgi. Moreover, vesicles mediate selective transfer of contents between Golgi cisternae. Thus, providing spatial cues to vesicles arriving at the Golgi is critical for endomembrane traffic and represents an ideal system to investigate the basis of vesicle targeting specificity.
One set of candidate vesicle tethers at the Golgi is the “golgins”, large well conserved coiled-coil proteins anchored to the membrane via their C-termini (9-11). The golgins have been suggested to act as tethers but in vivo evidence for this role is lacking (12, 13). In addition, other roles have been proposed for some golgins include recruiting kinases or cytoskeletal regulators (14, 15). Golgin mutants typically have mild or tissue-specific phenotypes, leaving their function uncertain (16, 17). We hypothesized that the golgins have overlapping functions in tethering, obscuring their function when any one is missing. We therefore developed methods to test their sufficiency, rather than necessity, in selective vesicle tethering.
Experimental strategy
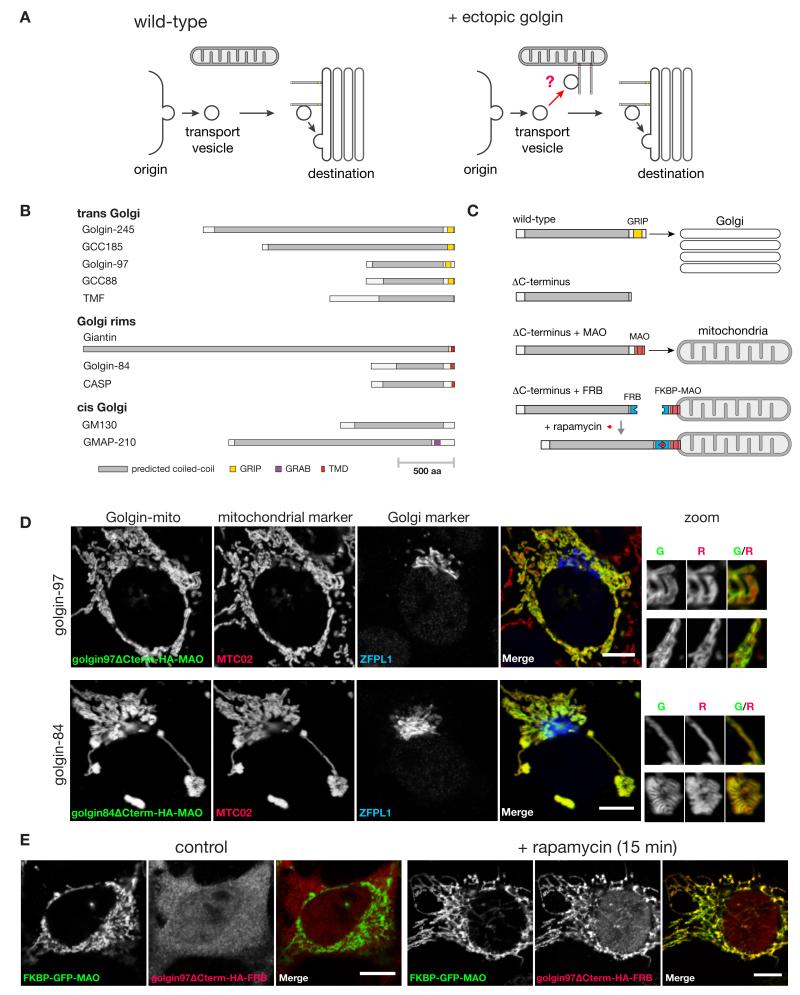
Our strategy was to attach golgins to a different structure in the cell and test if this was sufficient to redirect Golgi-bound carriers to the ectopic destination (Fig. 1A). We chose mitochondria because they do not normally capture transport vesicles, display a distinctive localization, and are distributed throughout the cell. We tested ten mammalian golgins chosen for their conservation outside of vertebrates and spatial distribution in different Golgi regions (Fig. 1B). Ectopic golgin localization was accomplished by either direct or induced attachment to a mitochondrial transmembrane domain in place of the C-terminal Golgi targeting domain (Fig. 1C). The direct fusion approach was more straightforward (Fig. 1D, and fig. S1), while the induced targeting strategy provided tight temporal control (Fig. 1, C and E). The golgins GM130 and GCC185 are known to bind soluble cytosolic proteins (p115 and CLASP respectively (14, 18)), and in both cases relocation of the golgin resulted in the binding partner being found on the mitochondria as well as the Golgi, suggesting that relocation had not grossly perturbed golgin structure (fig. S2). For two of the golgins (GM130 and GMAP-210), high level expression of the mitochondrial form caused the Golgi to become fragmented, which could either be due to their titrating away factors such as p115 that contribute to Golgi integrity, or to the effect of their relocation on ER to Golgi traffic (discussed further below).
Fig. 1. Strategy for relocation of the golgins to mitochondria.
(A) An in vivo assay for golgin tethering activity. Normally vesicles from various origins move to find the Golgi. If a particular class of vesicles can be captured by a specific golgin, then presenting that golgin on mitochondria should allow ectopic capture of that class of vesicle. (B) The ten human golgin coiled-coil proteins that have orthologs outside of vertebrates. (C) Scheme for relocating golgins to mitochondria in a constitutive or inducible manner, illustrated for the GRIP domain protein golgin-97. The C-terminal transmembrane domain of monoamine oxidase (MOA) is sufficient for targeting to the outer mitochondrial membrane (40). Inducible relocation was performed using the rapamycin-controlled hetero-dimerization domains from FKBP and FRB (41). (D) COS cells expressing representative golgin-mito fusion proteins stained for an HA epitope located between the golgin and the organelle targeting motif. Costaining for a mitochondrial marker, MTC02, and a Golgi marker, ZFPL1 (cis), indicate correct relocation of each of the golgins to the outer membrane of the mitochondria. Representative regions of mitochondria are magnified (Golgi marker omitted; therefore showing green and red channels only). Scale bars 10 μm. (E) COS cells coexpressing reroutable golgin-97 with FKBP-GFP-MAO with or without 200 nm rapamycin treatment for 15 minutes. Reroutable golgin-97 was detected by staining for an HA epitope tag between the golgin and the FRB domain. Scale bars 10 μm.
Vesicles originating from different locations were analysed for localization upon ectopic expression of individual golgins. The identity of originating vesicles was marked by either endogenous or exogenous cargo selective to that site. Analysis of all combinations of originating vesicles with each of the ten golgins was anticipated to provide a comprehensive view of their capacity to provide landmarks for vesicular traffic. We present these findings by successively analysing transport vesicles of endosomal, ER, and intra-Golgi origin.
Capture of endosome to Golgi carriers
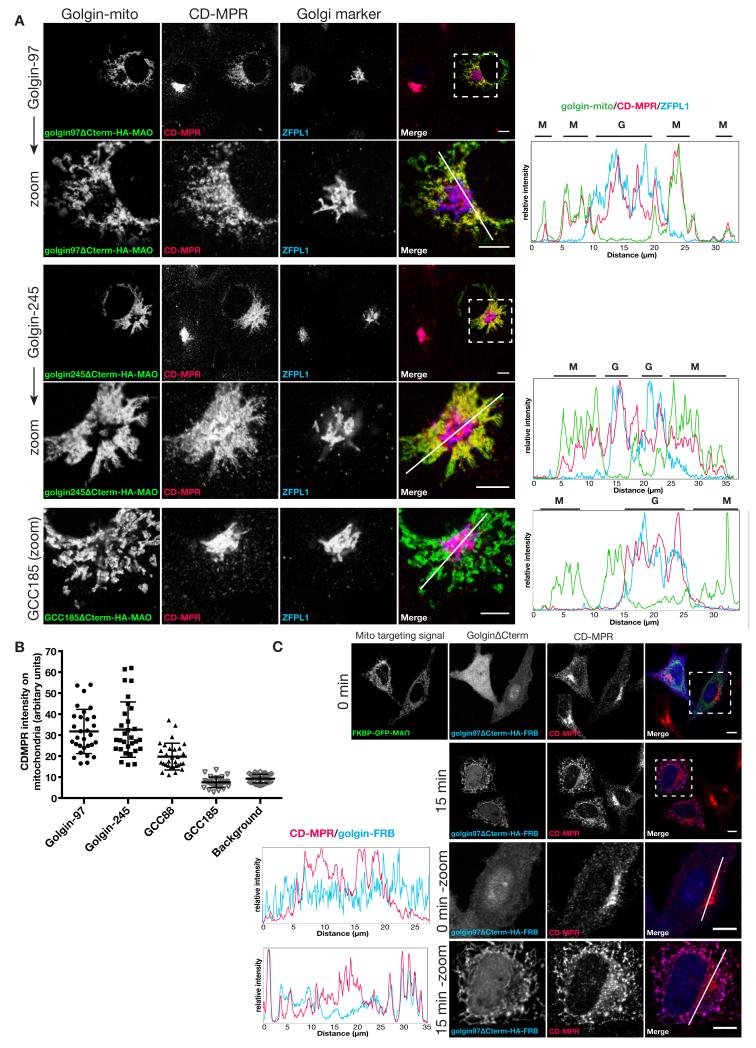
One of the best characterized cargos that traffics from endosomes back to the Golgi is the cation-dependent mannose 6-phosphate receptor (CD-MPR) that delivers newly made lysosomal hydrolases to endosomes and then returns to the Golgi (19). At steady state most of this receptor is in the Golgi apparatus, but mitochondrial forms of three of the golgins caused endogenous CD-MPR to accumulate on mitochondria (Fig. 2A). These were golgin-97, golgin-245 and GCC88, three of the four golgins that share a C-terminal GRIP domain that mediates binding to Arl1, a G protein of the trans Golgi (9, 11). In each case CD-MPR distribution was the sum of the Golgi and mitochondrial distribution, and quantitation confirmed that the effect was seen in most, if not all, of the transfected cells (Fig. 2B). The remaining seven golgins had no detectable effect on the distribution of the CD-MPR (Fig. 1A). The same three golgins could also induce a redistribution of the SNARE protein Vti1a, another cargo of these retrograde carriers (20) (fig. S3). Other endogenous proteins that travel along this retrograde route were also affected including the cation-independent MPR and TGN46, a protein of unknown function that has both Golgi retention and endosome retrieval signals (21) (fig. S4). These effects did not simply reflect capture of endosomes on the mitochondria because both early and late endosomal markers did not relocate (fig. S5). Finally, to confirm that the carriers were moving from endosomes to Golgi, we expressed the golgins in a cell line expressing a CD8-MPR chimera that recycles efficiently from the surface to the Golgi via endosomes (22). When CD8 antibodies were bound to surface and followed after endocytosis, the carriers containing the antibodies could be captured by the mitochondrial golgins early after initiation of uptake indicating that the carriers were moving from endosomes to the Golgi (fig. S6).
Fig. 2. Golgin-97, golgin-245 and GCC88 relocate endosome-to-TGN cargos to the mitochondria.
(A) COS cells expressing the indicated golgin-mito (mitochondria-targeted golgins) were costained for an HA epitope to detect the recombinant golgins, endogenous CDMPR, and ZFPL1 (cis) to mark the Golgi. Representative regions are shown magnified. Intensity plots of signal intensity (y-axis) against distance in μm (x-axis) show occurrence of overlap between channels. Respective signals from the mitochondria and the Golgi are indicated above each plot by letters M (for mitochondria) and G (for Golgi). (B) Plot for mean CDMPR labelling intensity within the mitochondria segment (the golgins) or elsewhere in the cell (background labelling). n ≥ 30. (C) COS cells coexpressing reroutable golgin-97 with FKBP-GFP-MAO were treated with 200 nm rapamycin for 0 or 15 minutes and costained for endogenous CDMPR. Intensity plot as in (A). Scale bars 10 μm.
The above experiments used golgins that were constitutively targeted to mitochondria in transiently-transfected cells, and so the golgins required 24-36 hours to accumulate to readily detectable levels. When the golgins were instead acutely located to mitochondria using the rapamycin-dependent inducible system we observed a rapid relocation of the retrograde cargo such as CD-MPR and Vti1a within 15 minutes of addition of rapamycin (Fig. 2C, and fig. S3B). This relocation was seen with the same three GRIP domain golgins, and suggests that the effect is a direct consequence of the ectopic location of the golgins (fig. S7).
Capture of ER to Golgi carriers
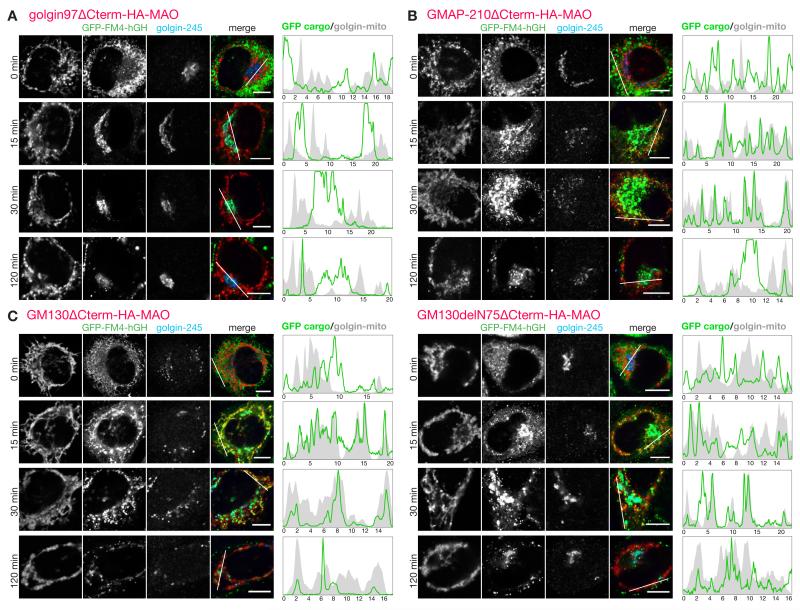
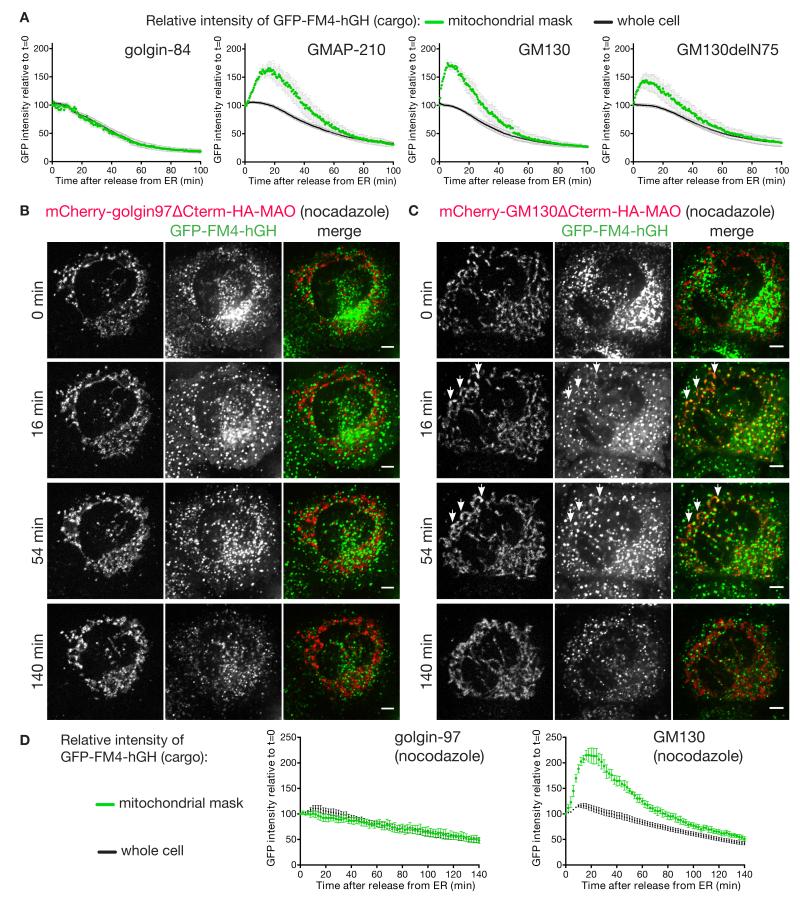
A second major class of carriers that arrive at the Golgi are those that deliver newly made secreted and membrane proteins from the ER. Proteins leave the ER from exit sites found both adjacent to the Golgi and scattered throughout the cytoplasm, and from the latter sites carriers move along microtubules to reach the Golgi (23). To follow these carriers we made use of a GFP-tagged secreted protein that accumulates in the ER until released by a small molecule (24). Upon release the reporter relocates to the Golgi within 10-15 minutes, and then leaves for the surface, and this behaviour was not perturbed by the mitochondrial forms of eight of the ten golgins tested (Fig. 3A). However for the golgins GM130 and GMAP-210 we observed association of GFP-labelled structures with mitochondria (Fig. 3, B and C). This association appeared to gradually reduce after an initial peak, and to quantify this we used live cell imaging to compare the GFP-labelled cargo to mCherry-labelled mitochondrial golgins in individual cells over time (fig. S8-S11). Quantification revealed that GM130 and GMAP-210 caused an association of ER-derived carriers with mitochondria that was specific but also transient, presumably reflecting the carriers being captured and then eventually released, perhaps by being pulled away by the motors that move them toward the Golgi (Fig. 4A).
Fig.3. GMAP210 and GM130 capture ER-derived cargo on to mitochondria.
(A-C) C1-HeLa cells expressing GFP-FM4-hGH cargo and transiently expressing mitochondria-targeted golgin-97 (A), GMAP210 (B), GM130 (C) or GM130ΔN75 (C) were subjected to a secretion assay by treatment with 0.5 μM D/D Solubilizer at 37°C and fixed at the indicated time points. The number 75 in GM130ΔN75 represents the number of the first residue in the fragment. Transfected C1 cells were permeabilized and costained for an HA epitope, to detect the recombinant golgins; and a Golgi marker, golgin-245 (trans). Intensity plots of signal intensity (y-axis) against distance in μm (x-axis) show occurrence of overlap between channels. Scale bars 10 μm.
Fig.4. Live cell imaging of ectopic capture of ER-derived carriers by GMAP210 and GM130.
(A) Graphs of GFP fluorescence (y-axis) against time (x-axis) demonstrate time-courses for secretion of the GFP-FM4-hGH reporter by C1 cells that were transiently expressing mitochondria-targeted golgins. Average GFP fluorescence intensity over the entire cell (whole cell) or the mitochondrial region (mitochondria) was determined for several cells over time, with the results presented as percentages of the fluorescence intensity at t=0. Data are mean +/− SEM; golgin-84 (N=5), GMAP-210 (N=7), GM130 (N=6), and GM130delN75 (N=7). Stills from a representative data set from each are shown in fig. S8-S11. (B,C) C1-HeLa cells transiently expressing mitochondria-targeted mCherry-golgin-97 (B), or mCherry-GM130 (C) were pre-treated with 0.5 μM nocodazole for three hours to depolymerize microtubules prior to being incubated at 37°C for the indicated times with a mix of nocodazole and D/D solubilizer to induce secretion of the GFP-FM4-hGH reporter. Images were acquired every 2 minutes for 140 minutes (movies S1 and S2). Transient association of GFP-FM4-hGH with mitochondria can be seen for GM130 (illustrated by arrowheads), but not golgin-97. Scale bars 5 μm. (D) Quantitation of the relative association of the GFP-FM4-hGH reporter with mitochondria over time in nocodazole treated cells as in B and C. Data are mean +/− SEM (N=4 for both).
The two golgins able to capture pre-Golgi carriers are both found on the cis side of the Golgi, consistent with a role in ER to Golgi traffic (25, 26). GM130 interacts via its N-terminal 75 residues with the tethering factor p115 (18). A mitochondrial form of GM130 lacking residues 1-75 no longer relocated p115 to mitochondria and showed reduced Golgi fragmentation (Fig. 3C, fig. S2). Nonetheless, this construct could still transiently capture ER-derived carriers suggesting that GM130 has some vesicle tethering activity independent of p115, and also indicating that the ectopic capture of ER-derived cargo is not simply a consequence of Golgi fragmentation (Fig. 3C and 4A). To strengthen the latter conclusion we also followed ER-derived cargo in cells in which the Golgi had been fragmented by depolymerizing microtubules. This showed that capture was still not observed with a control golgin, but could still be clearly detected for GM130 (Fig. 4, B to D, movies S1 and S2).
Capture of carriers containing Golgi residents
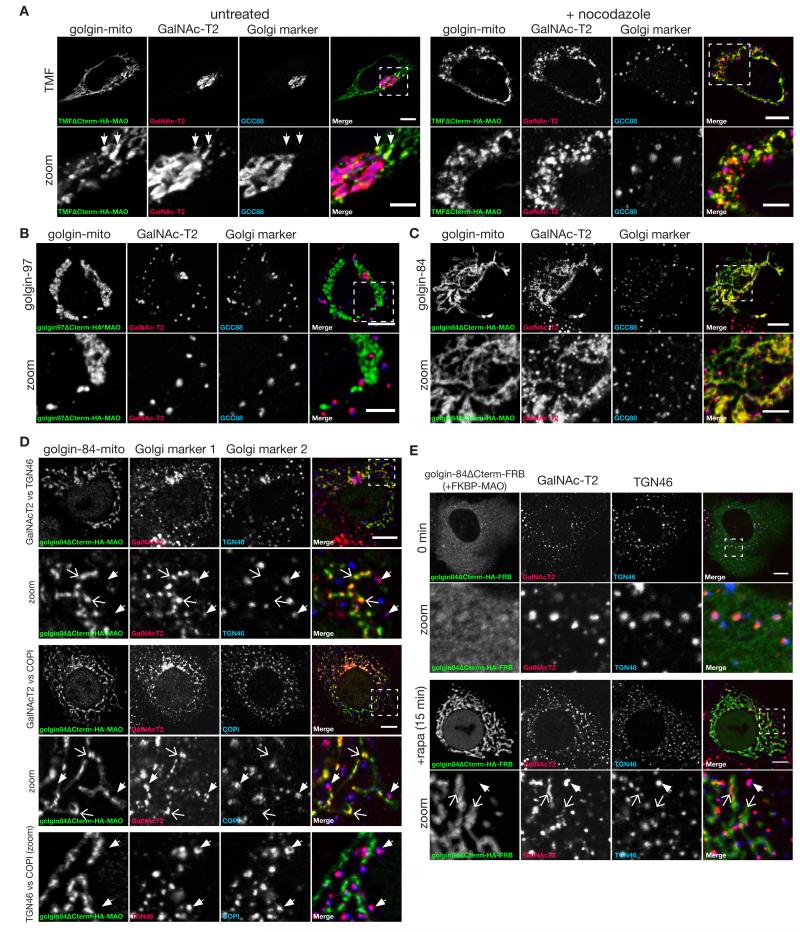
The Golgi stack is surrounded by vesicles that are thought to recycle Golgi resident proteins back to earlier parts of the stack as the cisternae mature, and it has also been proposed that some may carry secretory cargo forward through the stack (27). We thus looked for relocation of Golgi resident proteins by the relocated golgins. With ectopic TMF the endogenous Golgi enzyme GalNAc-T2 could be detected on mitochondria as well as on the Golgi but, unlike the carriers described above, this accumulation was only seen on those mitochondria near to the Golgi stack (Fig. 5A). This may reflect the fact that intra-Golgi carriers do not need to move far from the Golgi and so most only encounter the few mitochondria that are close to the stack, whereas carriers coming to the Golgi from the ER and endosomes can travel long distances along microtubules and hence pass many mitochondria. To increase the probability of intra-Golgi carriers encountering mitochondria we used nocodazole to remove microtubules and thus scatter the Golgi into many mini-stacks that are located throughout the cytoplasm and so in the proximity of many more mitochondria. In such cells we could see robust relocation of GalNAc-T2 to mitochondria by TMF (Fig. 5A), and also by golgin-84 and GMAP-210 but not any other golgin (Fig. 5, B and C; fig. S12). Analysis of further Golgi markers confirmed the mini-stacks are distinct from the GalNAc-T2 captured on mitochondria because the markers from different cisternae are much closer to each other than to the GalNAc-T2 on the mitochondria (Fig. 5D). Finally, following acute relocalization of golgin-84 to mitochondria using the rapamycin-based dimerization GalNAc-T2 could be clearly detected on mitochondria after 15 minutes, indicating that this capture in nocadazole-treated cells is unlikely to be due to an indirect effect arising over many hours (Fig. 5E).
Fig. 5. Mitochondrial TMF and golgin-84 relocate Golgi membrane proteins.
(A) Confocal micrographs of COS cells expressing TMF-mito (HA) and stained for a Golgi enzyme, GalNAc-T2 and a Golgi marker, GCC88 (trans), without, or following, six hours nocodazole treatment as indicated. (B,C) COS cells expressing the indicated golgin-mito were treated with nocodazole for 6 hours and costained for GalNAc-T2 and a Golgi marker, GCC88 (trans). (D) COS cells expressing golgin-84-mito were treated with 0.5 μM nocodazole for 6 hours and labelled for two of the endogenous Golgi proteins GalNAcT2, TGN46 or COPI as indicated. In all cases, closed arrows indicate Golgi ministacks positive for both Golgi markers, with these being distinct from mitochondria, whilst open arrows indicate structures on mitochondria positive for GalNAcT2 but not the other marker. (E) COS cells coexpressing golgin-84ΔCterm-FRB and FKBP-MAO were treated with 0.5 μM nocodazole for three hours followed by 200 nm rapamycin (and nocadazole) direct dimerization, and then fixed as indicated and stained for endogenous GalNAcT2 and TGN46. Closed arrows indicate Golgi ministacks positive for both markers, and open arrows GalNAcT2 captured onto mitochondria after 15 minutes. Scale bars 10 μm.
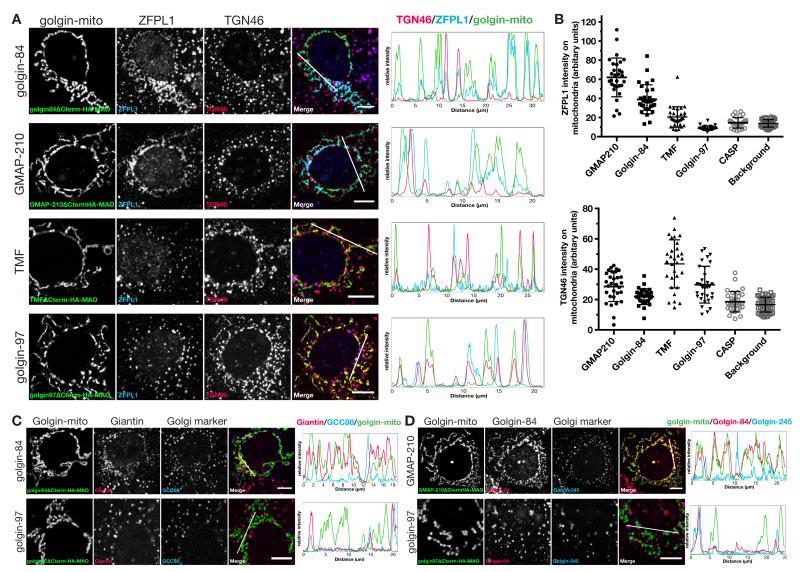
We next examined a range of different Golgi resident membrane proteins in nocodazole-treated cells and again found relocation by specific golgins, with strikingly different patterns seen for different residents. The cis Golgi membrane protein ZFPL1 (28), was efficiently relocated by golgin-84 and GMAP-210, and to a lesser extent by TMF, but unaffected by the other seven golgins (Fig. 6A and fig. S13). In contrast the TGN resident protein TGN46 was relocated by TMF, but not by golgin-84 or GMAP-210 (Fig. 6A). Quantitation of colocalization with mitochondria confirmed these differential relocations (Fig. 6B). TGN46 has two targeting signals, one in its cytoplasmic tail which mediates retrieval from endosomes (and hence TGN46 is in the endosome to Golgi carriers captured by the GRIP golgins), and a second transmembrane signal that confers retention in the trans-Golgi, probably by directing recycling within the stack (21).
Fig. 6. TMF, golgin-84 and GMAP210 exhibit specificity in the capture of cis-Golgi and trans-Golgi residents.
(A) COS cells expressing the indicated golgin-mito were treated with nocodazole for 6 hours and costained for two transmembrane Golgi markers, ZFPL1 (cis) and TGN46 (trans). Intensity plots of signal intensity (y-axis) against distance in μm (x-axis) show occurrence of overlap between channels. (B) Quantitation of mean intensity of ZFPL1 signal and TGN46 signal within the HA-positive mitochondria segment (the golgins) or whole cell (background), n ≥ 30. (C,D) COS cells expressing the indicated golgin-mito were treated with nocodazole for 6 hours and costained for either of the transmembrane golgins, giantin or golgin-84, and a Golgi marker, GCC88. Intensity plots of signal intensity (y-axis) against distance in μm (x-axis) show occurrence of overlap between the markers. Scale bars 10 μm or 5 μm (magnified boxes).
The golgins giantin, CASP and golgin-84 are themselves Golgi resident membrane proteins and so if Golgi cisternae undergo maturation they would have to recycle to earlier cisternae in transport vesicles. Indeed, endogenous giantin was relocated to mitochondria by golgin-84 and GMAP-210, and although golgin-84 could not be tested with itself, it was also relocated by GMAP-210 (Fig. 6, C and D; and fig. S14). It is possible that these golgins interact directly to mediate tethering, but this remains to be determined. Taken together these results show that three of the golgins are able to relocate resident membrane proteins of the Golgi to mitochondria. This presumably reflects the capture of carrier vesicles containing these proteins, with the pattern of proteins captured varying between individual golgins.
Electron microscopy reveals carrier capture by golgin-coated mitochondria
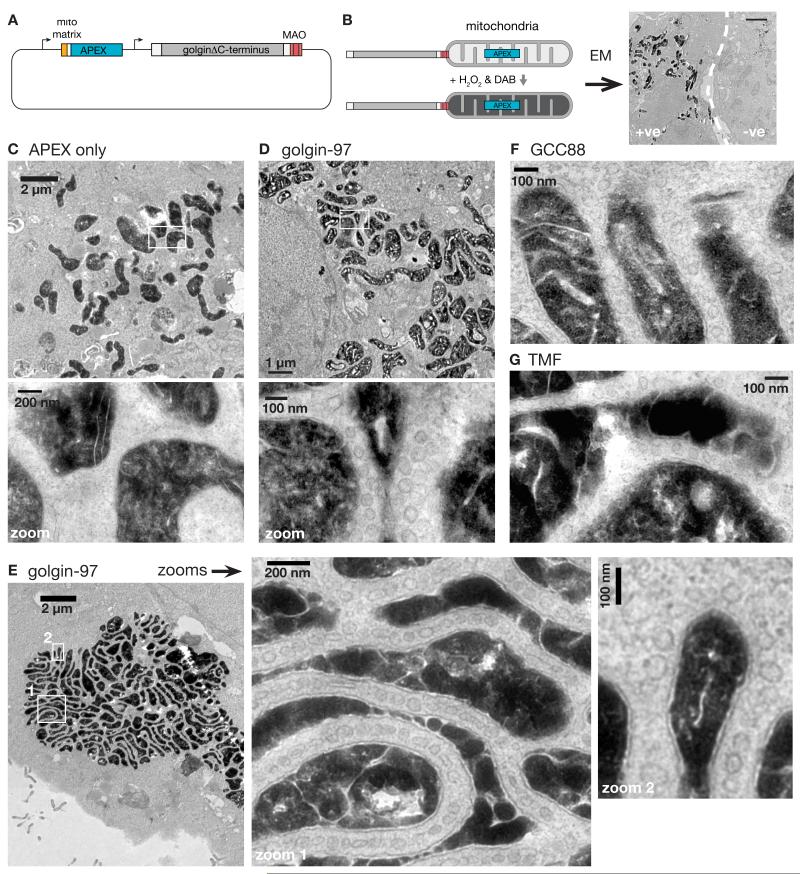
In the above experiments endogenous membrane proteins were found to be relocated to mitochondria by specific golgins. This predicted that membrane-bound carriers should be seen accumulating around mitochondria, and to investigate this we used electron microscopy (EM). To identify transfected cells the mitochondrial golgins were expressed from a bi-cistronic plasmid that also expressed a form of APEX peroxidase that is targeted to the mitochondrial matrix (Fig. 7A). This peroxidase directs the precipitation of diaminobenzidine to form a dark deposit that is visible in EM sections (29) (Fig. 7B). We initially examined the effect of co-expressing golgin-97 which we had found to efficiently capture endosome derived-proteins. In these cells the mitochondria were surrounded by circular and oval membrane profiles that were absent from control cells expressing APEX alone (Fig. 7, C and D). The structures were typically 60-80 nm in diameter and often accumulated between adjacent mitochondria, but could also be seen around isolated parts of the mitochondrial surface.
Fig. 7. Ultrastructural evidence of golgins as vesicle tethers and model for golgin function.
(A) Plasmid for coexpression of mitochondrial-matrix-targeted APEX and golgin-mito. (B) Stratagey for identifying cells labelled by APEX-catalysed oxidation of DAB. Scale bar 2 μm. (C) Electron micrograph of a COS cell transfected with a plasmid expressing mito matrix-APEX alone. (D-G) Electron micrographs of COS cells coexpressing mito matrix-APEX and either golgin-97-mito (D,E), GCC88-mito (F), or TMF-mito (G). In all cases membranous vesicles can be seen accumulating around the mitochondria.
Such membrane capture was also observed with other golgins that can relocate membrane proteins, including GCC88, TMF, GMAP-210, golgin-245 and golgin-84 (Fig. 7, F and G; and fig. S15). In contrast we did not observe membranes accumulating around mitochondria coated with giantin or GCC185, consistent with their lack of effect on any of the markers tested (fig. S16). The tendency for the captured tubulovesicular structures to accumulate between mitochondria is presumably due to them simultaneously engaging golgins attached to adjacent mitochondria. This “zippering” effect resulted in clustering of mitochondria, and explains the striped appearance of golgins on mitochondrial clusters that we had observed by immunofluorescence (Fig. 1D). In some cells, the mitochondrial clustering was so effective that all were collected together, separated by striking lines of captured vesicles that were often at a regular spacing between the mitochondria (Fig. 7E).
Discussion
Multiple classes of intracellular transport vesicles deliver cargo specifically to the Golgi apparatus. Here we show that relocation of specific golgin coiled-coil proteins is sufficient to redirect these specific classes of vesicles to an ectopic location (see Figure 8A for a summary). This provides clear in vivo evidence for at least seven of the golgins being bona fide vesicle tethers. However, the golgins not only capture vesicles, but they can clearly distinguish vesicles from different origins. This implies that the location of golgins specifies the point of vesicle tethering, and indicates that the machinery that recruits the golgins to specific cisternae of the Golgi helps to define their identity as the correct target organelle for incoming vesicles (Fig. 8B). This does not preclude other factors such as SNARE complex formation also making a contribution to the specificity of membrane traffic but, at least in this case, SNARE complex formation does not appear to be necessary for recognition of the destination organelle.
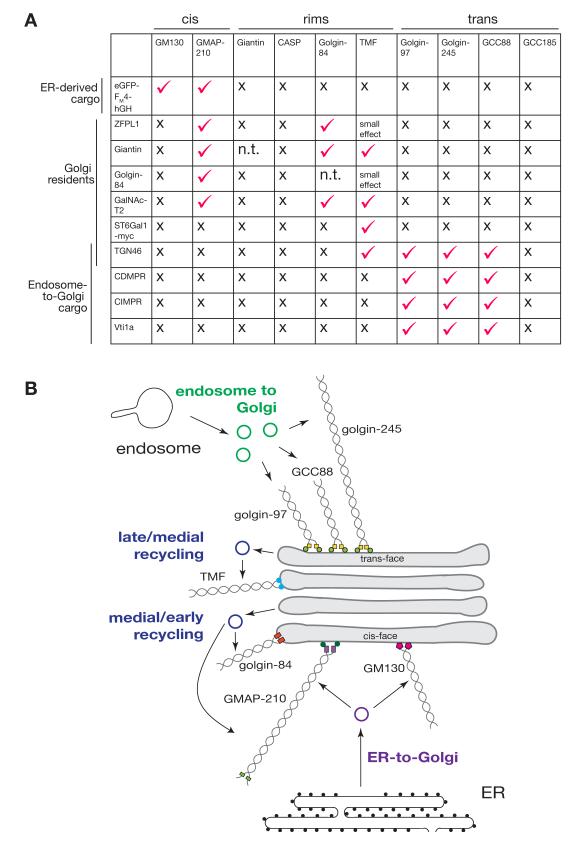
Fig. 8. Summary of golgin tethering activities.
(A) Summary of results of relocation of the indicate membrane proteins by the panel of ten golgins that tested. Tick indicates capture at the mitochondria in the majority of cell examined, x indicates absence of detectable effect on intracellular distribution. (B) Schematic of how golgins appear to tether the different sets of vesicles arriving at the Golgi. Golgin-97, golgin-245 and GCC88 capture endosome-to-TGN carriers; TMF, golgin-84 and GMAP210 mediate intra-Golgi transport, with the latter two having a preference for early Golgi residents; GMAP210 and GM130 capture ER-to-Golgi carriers.
Our studies also reveal a degree of redundancy within the golgins, with there being subsets that apparently share tethering activity. For instance golgin-97, golgin-245 and GCC88 all capture endosome to Golgi cargo, whilst GM130 and GMAP-210 capture ER to Golgi carriers. This partial redundancy would be consistent with the proposal that they act collectively to surround different regions of the Golgi with docking sites for particular vesicle types (30). Such a model would be loosely analogous to the capture of cytoplasmic to nuclear cargo by the multiple FG repeat proteins of the nuclear pore (31). Three of the ten golgins (CASP, GCC185 and giantin) did not show tethering activity in our assay and so may have other roles such as that of organising microtubules as has been suggested for GCC185 (14). However, it is also possible that they can act as tethers, but only in conjunction with other golgins, or following Golgi-specific modification (32, 33).
The fact that golgins can distinguish different classes of carrier vesicle implies that each class displays one or more unique feature that allow it to be recognized. There are a range of plausible candidates for these vesicle features such as the Rab GTPases (30, 34, 35). Indeed further proteins may bind directly to the golgins to help capture the vesicle even if the golgins are themselves sufficient to nucleate a functional tethering entity. However, irrespective of what else is involved in completing the entire tether, our findings already open up routes to identify and isolate specific classes of carrier vesicles. These key mediators of traffic are small and normally short-lived, but the ectopic golgins cause these carriers to accumulate in a docked state which allows their contents to be readily probed by immunolabeling, and also suggests routes to isolation for proteomic analysis. In the case of the Golgi our studies are already informative with trans Golgi residents preferring TMF, whilst those from the cis-and medial-Golgi prefer golgin-84 and GMAP-210. Since the golgins are presented in the same way (ie on mitochondria adjacent to the Golgi), this finding indicates that the vesicles from different parts of the stack must display different golgin-binding features, and hence their destination within the stack is directed at least in part by molecular interactions and not simply by spatial proximity. This may be important for allowing the structure of the Golgi to vary between tissues and yet still maintain cisternal identity. In addition, the finding that GMAP-210 can bind both ER-derived vesicles and those containing Golgi residents, raises the intriguing possibility that it could help to scaffold the assembly of new cisternae. As such these findings not only reveal that golgins are sufficient to confer specificity to the tethering of Golgi destined vesicles, but also provide new tools to investigate the organisation of membrane traffic in this complex and much debated organelle (36).
Materials and Methods
Plasmids
C-terminally truncated golgins were provided by Alison Gillingham (human GMAP210, golgin-84 and CASP (33, 37)), or PCR amplified from full-length cDNAs kindly provided by others (human GCC88, GCC185 and golgin-245 from Paul Gleeson (38); human golgin-97 from Francis Barr; mouse TMF and rat GM130 from Martin Lowe; human giantin from Adam Linstedt (39)). Human MAO-A C-terminal TMD (481-528) was PCR amplified from human cDNA.
Truncated golgins were as follows: GCC88ΔC-term (1-Ala718); GCC185ΔC-term (1-Ser1535); golgin-97ΔC-term (1-Val681); golgin-245ΔC-term (1-Gly2163); TMFΔC-term (1-Thr779); giantinΔC-term (1-Cys3213); golgin-84ΔC-term (1-Ala677); CASPΔC-term (1-Arg618); GM130ΔC-term (1-Leu872); GM130ΔN75ΔC-term (Met75-Leu872); GMAP-210ΔC-term (1-Leu1756).
GolginΔC-term-GAGAGA linker-HA-MAO, golginΔC-term-GAGAGA(or GAGAGS) linker-HA-FRB, mCherry-GAGAGA(or GSGSGS) linker-golginΔC-term-GAGAGA linker-MAO, FKBP-GFP-MAO and FKBP-flag-MAO were cloned into COS cell expression vector containing the cytomegalovirus promoter. Bicistronic plasmids for coexpression of mito-APEX (Addgene #42607; (29)).and golginΔC-term-GAGAGA linker-HA-MAO were generated in pcDNA3.1+ (Clontech) from the respective monocistronic plasmids.
Antibodies
Antibodies used in this study include: mouse CD-MPR (22d4, Developmental Studies Hybridoma Bank (DSHB)); mouse CD8 (UCHT4, Sigma); rabbit CLASP1 (gift from Anna Akhamanova); rat β′COP (23C (TCP-1)); mouse EEA1 (610457, BD Biosciences); mouse ERGIC-53 (ALX-804-602-C100, Enzo Life Sciences); mouse GalNAc-T2 (UH-4, gift from Henrik Clausen), rabbit GCC88 (HPA021323, Sigma); rabbit GCC185 (HPA035849, Sigma); rabbit giantin (HPA011008, Sigma); rabbit GM130 (1837-1, Epitomics); mouse GM130 (610823, BD Biosciences); mouse golgin-245 (611281, BD Biosciences); rabbit golgin-84 (HPA000992, Sigma); rabbit HA (Y-11, Santa Cruz Biotechnology); rat HA (3F10, Roche); mouse LAMP1 (H4A3, DSHB); mouse MTC02 (ab3298, Abcam); mouse p115 (612260, BD Biosciences); sheep TGN46 (AHP500, AbD serotec); rabbit TMF (HPA008729, Sigma); mouse Vti1a (611220, BD Biosciences); rabbit ZFPL1 (HPA014909, Sigma).
Cell culture, transient transfections and treatments
COS-7 cells, CD8-CIMPR HeLa cells (22), C1-HeLa cells expressing eGPF-FM4-hGH (24), and mouse embryonic fibroblasts (MEFs) were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Invitrogen) supplemented with 10% fetal calf serum (FCS) and penicillin/streptomycin at 37°C and 5% CO2.
COS-7 cells and MEFs were transiently transfected with Fugene6 (Promega), and HeLa cell lines with Xtreme Gene 9 (Roche) according to the manufacturers’ recommendations. Cells plated on 6-well plates were transfected 10-18 hours after plating when cells had reached 50%-80% confluency. After the transfection mixtures were applied, cells were analysed 24-48 hours post-transfection.
To induce heterodimerization of FKBP and FRB domains, cells co-expressing fusion proteins with each of these domains were treated at 37°C for various incubation times with rapamycin (Sigma; 200 nm from 2.74 mM stock in DMSO). To depolymerize microtubules, cells were treated at 37°C for 2-6 hours with nocodazole (Sigma, 1 μm from a 1 mM stock in DMSO). To induce secretion in C1-HeLa cells, cells were treated with D/D solubilizer (Clontech, 0.5 μM) at 37°C for various incubation times (24).
HeLa cells stably expressing the CD8-CIMPR reporter construct (22), were transfected with golgin-mito constructs 24-48 hours before the internalisation assay and plated onto multispot microscope slides 12-36 hours post-transfection. Slides were incubated at 4°C for 20 minutes to stop all trafficking and washed with PBS. Mouse anti-CD8 antibody (Sigma) was applied in DMEM for 30 minutes at 4°C to bind onto the cell surface. Following two washes with PBS, antibody uptake was induced by replacing PBS with pre-warmed DMEM followed by incubation at 37°C prior to fixation with 4% formaldehyde in PBS.
Immunofluorescence and live cell imaging
COS-7 cells, HeLa cells and MEFs were fixed with 4% formaldehyde in PBS for 30 minutes, permeabilized in 0.5% TritonX-100 for 10 minutes, and blocked in blocking buffer (20% FCS, 0.5% tween-20 in PBS) for 30 minutes. Primary and secondary antibodies were applied in blocking buffer for 1 hour, cells were washed twice with PBS and mounted under a coverslip in Vectashield mounting medium (Vector Labs). Images were acquired with a Zeiss LSM780 confocal microscope using a 63× Apochromat oil-immersion objective.
Quantitation of mitochondrial capture in fixed cells was performed with Imaris 7.4.0. (Bitplane). First, the mitochondria and Golgi regions of each transfected cell were segmented by the HA staining pattern and ZFPL1 staining respectively. Next, the latter segment was subtracted from the former segment to create a new mitochondria segment which had excluded any overlapping Golgi regions. Mean intensity of the molecule of interest on this segment were determined as an arbitrary, but relative, value. Background staining levels were obtained by quantifying mean signal levels on a whole cell segment subtracting the Golgi segment.
HeLa cells stably expressing GFP-FM4-hGH (C1-HeLa, (24)) were seeded onto 4-well chambers (Lab-Tek) in complete medium. Prior to imaging, 10 mM HEPES (Sigma) was applied to the cells. Live cells were imaged at 37°C using a 100× oil objective on a Nikon Eclipse Ti inverted microscope equipped with an Andor Revolution XD system and a Yokogawa CSU-X1 and an Andor iXon EMCCD camera. Quantitation of transient association of GFP-FM4-hGH with mitochondria decorated with golgins was as for fixed cells, except the mitochondria were segmented using the mCherry fluorescence. At each time point of a movie, two sets of mean signal intensity of the molecule of interest were obtained; one from the mitochondria segment and the other from the whole cell segment.
APEX and electron microscopy
Cells grown on glass bottom petri dishes (MatTek) were fixed at room temperature with 2% glutaraldehyde (EM grade, Agar Scientific) in 0.1 M cacodylate buffer pH 7.4 (CB). Cells were then incubated on ice for 30-60 min, rinsed twice with chilled CB, and blocked with 50 mM glycine in CB on ice for 10 minutes. To start the APEX-catalysed oxidation of 3,3-diaminobenzidine (DAB), a freshly prepared solution of 0.5 mg/mL DAB free base (Sigma) in HCL and 0.03% (10 mM; Sigma) H2O2 were added to the cells. Formation of reaction product (a brownish precipitate) was monitored by light microscopy. After 15 minutes, the reaction was halted by washing the cells twice with chilled CB. Samples were post-fixed with 1% osmium tetroxide in CB on ice for 1 hour, washed five times with distilled water, and dehydrated in an ascending ethanol series (30%, 50%, 70%, 90%, 100%) at room temperature. The dehydrated samples were infiltrated and embedded in CY212 resin. Areas containing brown precipitates were excised and mounted on dummy blocks and sectioned parallel to the substratum. Pale gold, 70 nm sections were contrasted with saturated aqueous uranyl acetate and Reynolds lead citrate. Electron micrographs were recorded at 80 kV or 120 kV on a FEI Tecnai Spirit TEM.
Supplementary Material
Acknowledgements
We are indebted to Anna Akhmanova, Francis Barr, Henrik Clausen, Paul Gleeson, Adam Linstedt, Martin Lowe, Ulla Mandel, Margaret Robinson and Matthew Seaman for generous provision of reagents, and to Gillian Howard for help with electron microscopy. We thank Alison Gillingham, Ramanujan Hegde, Ben Nichols and Katja Röper for comments on the manuscript. Funding was from the UK Medical Research Council (MRC file reference number U105178783). The data are presented in the main manuscript and the supplementary materials.
References and Notes
- 1.Jahn R, Scheller RH. SNAREs - engines for membrane fusion. Nat. Rev. Mol. Cell Biol. 2006;7:631–643. doi: 10.1038/nrm2002. [DOI] [PubMed] [Google Scholar]
- 2.Söllner T, et al. SNAP receptors implicated in vesicle targeting and fusion. Nature. 1993;362:318–324. doi: 10.1038/362318a0. [DOI] [PubMed] [Google Scholar]
- 3.McNew JA, et al. Compartmental specificity of cellular membrane fusion encoded in SNARE proteins. Nature. 2000;407:153–159. doi: 10.1038/35025000. [DOI] [PubMed] [Google Scholar]
- 4.Willett R, et al. COG complexes form spatial landmarks for distinct SNARE complexes. Nat. Commun. 2013;4:1553. doi: 10.1038/ncomms2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Whyte JRC, Munro S. Vesicle tethering complexes in membrane traffic. J Cell Sci. 2002;115:2627–2637. doi: 10.1242/jcs.115.13.2627. [DOI] [PubMed] [Google Scholar]
- 6.Waters MG, Pfeffer SR. Membrane tethering in intracellular transport. Curr Opin Cell Biol. 1999;11:453–459. doi: 10.1016/s0955-0674(99)80065-9. [DOI] [PubMed] [Google Scholar]
- 7.Yu I-M, Hughson FM. Tethering factors as organizers of intracellular vesicular traffic. Annu Rev Cell Dev Biol. 2010;26:137–156. doi: 10.1146/annurev.cellbio.042308.113327. [DOI] [PubMed] [Google Scholar]
- 8.Glick BS, Nakano A. Membrane traffic within the Golgi apparatus. Annu Rev Cell Dev Biol. 2009;25:113–132. doi: 10.1146/annurev.cellbio.24.110707.175421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goud B, Gleeson PA. TGN golgins, Rabs and cytoskeleton: regulating the Golgi trafficking highways. Trends Cell Biol. 2010;20:329–336. doi: 10.1016/j.tcb.2010.02.006. [DOI] [PubMed] [Google Scholar]
- 10.Ramirez IB, Lowe M. Golgins and GRASPs: holding the Golgi together. Semin. Cell Dev. Biol. 2009;20:770–779. doi: 10.1016/j.semcdb.2009.03.011. [DOI] [PubMed] [Google Scholar]
- 11.Munro S. The golgin coiled-coil proteins of the Golgi apparatus. Cold Spring Harb. Perspect. Biol. 2011;3 doi: 10.1101/cshperspect.a005256. doi:10.1101/cshperspect.a005256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Malsam J, Satoh A, Pelletier L, Warren G. Golgin tethers define subpopulations of COPI vesicles. Science. 2005;307:1095–1098. doi: 10.1126/science.1108061. [DOI] [PubMed] [Google Scholar]
- 13.Drin G, Morello V, Casella J-F, Gounon P, Antonny B. Asymmetric tethering of flat and curved lipid membranes by a golgin. Science. 2008;320:670–673. doi: 10.1126/science.1155821. [DOI] [PubMed] [Google Scholar]
- 14.Efimov A, et al. Asymmetric CLASP-dependent nucleation of noncentrosomal microtubules at the trans-Golgi network. Dev. Cell. 2007;12:917–930. doi: 10.1016/j.devcel.2007.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Preisinger C, et al. YSK1 is activated by the Golgi matrix protein GM130 and plays a role in cell migration through its substrate 14-3-3zeta. J. Cell Biol. 2004;164:1009–1020. doi: 10.1083/jcb.200310061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elkis Y, et al. Testosterone deficiency accompanied by testicular and epididymal abnormalities in TMF(−/−) mice. Mol. Cell. Endocrinol. 2013;365:52–63. doi: 10.1016/j.mce.2012.09.003. [DOI] [PubMed] [Google Scholar]
- 17.Smits P, et al. Lethal skeletal dysplasia in mice and humans lacking the golgin GMAP-210. N. Engl. J. Med. 2010;362:206–216. doi: 10.1056/NEJMoa0900158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakamura N, Lowe M, Levine TP, Rabouille C, Warren G. The vesicle docking protein p115 binds GM130, a cis-Golgi matrix protein, in a mitotically regulated manner. Cell. 1997;89:445–455. doi: 10.1016/s0092-8674(00)80225-1. [DOI] [PubMed] [Google Scholar]
- 19.Braulke T, Bonifacino JS. Sorting of lysosomal proteins. Biochim. Biophys. Acta. 2009;1793:605–614. doi: 10.1016/j.bbamcr.2008.10.016. [DOI] [PubMed] [Google Scholar]
- 20.Mallard F, et al. Early/recycling endosomes-to-TGN transport involves two SNARE complexes and a Rab6 isoform. J. Cell Biol. 2002;156:653–664. doi: 10.1083/jcb.200110081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ponnambalam S, Rabouille C, Luzio JP, Nilsson T, Warren G. The TGN38 glycoprotein contains two non-overlapping signals that mediate localization to the trans-Golgi network. J. Cell Biol. 1994;125:253–268. doi: 10.1083/jcb.125.2.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seaman MNJ. Cargo-selective endosomal sorting for retrieval to the Golgi requires retromer. J. Cell Biol. 2004;165:111–122. doi: 10.1083/jcb.200312034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Budnik A, Stephens DJ. ER exit sites--localization and control of COPII vesicle formation. FEBS Lett. 2009;583:3796–3803. doi: 10.1016/j.febslet.2009.10.038. [DOI] [PubMed] [Google Scholar]
- 24.Gordon DE, Bond LM, Sahlender DA, Peden AA. A targeted siRNA screen to identify SNAREs required for constitutive secretion in mammalian cells. Traffic. 2010;11:1191–1204. doi: 10.1111/j.1600-0854.2010.01087.x. [DOI] [PubMed] [Google Scholar]
- 25.Nakamura N, et al. Characterization of a cis-Golgi matrix protein, GM130. J. Cell Biol. 1995;131:1715–1726. doi: 10.1083/jcb.131.6.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Infante C, Ramos-Morales F, Fedriani C, Bornens M, Rios RM. GMAP-210, A cis-Golgi network-associated protein, is a minus end microtubule-binding protein. J. Cell Biol. 1999;145:83–98. doi: 10.1083/jcb.145.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Glick BS, Luini A. Models for Golgi traffic: a critical assessment. Cold Spring Harb. Perspect. Biol. 2011;3:a005215. doi: 10.1101/cshperspect.a005215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chiu C-F, et al. ZFPL1, a novel ring finger protein required for cis-Golgi integrity and efficient ER-to-Golgi transport. EMBO J. 2008;27:934–947. doi: 10.1038/emboj.2008.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martell JD, et al. Engineered ascorbate peroxidase as a genetically encoded reporter for electron microscopy. Nat. Biotechnol. 2012;30:1143–1148. doi: 10.1038/nbt.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sinka R, Gillingham AK, Kondylis V, Munro S. Golgi coiled-coil proteins contain multiple binding sites for Rab family G proteins. J. Cell Biol. 2008;183:607–615. doi: 10.1083/jcb.200808018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wälde S, Kehlenbach RH. The Part and the Whole: functions of nucleoporins in nucleocytoplasmic transport. Trends Cell Biol. 2010;20:461–469. doi: 10.1016/j.tcb.2010.05.001. [DOI] [PubMed] [Google Scholar]
- 32.Reddy JV, et al. A functional role for the GCC185 golgin in mannose 6-phosphate receptor recycling. Mol. Biol. Cell. 2006;17:4353–4363. doi: 10.1091/mbc.E06-02-0153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gillingham AK, Pfeifer AC, Munro S. CASP, the alternatively spliced product of the gene encoding the CCAAT-displacement protein transcription factor, is a Golgi membrane protein related to giantin. Mol. Biol. Cell. 2002;13:3761–3774. doi: 10.1091/mbc.E02-06-0349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Short B, Haas A, Barr FA. Golgins and GTPases, giving identity and structure to the Golgi apparatus. Biochim. Biophys. Acta. 2005;1744:383–395. doi: 10.1016/j.bbamcr.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 35.Satoh A, Wang Y, Malsam J, Beard MB, Warren G. Golgin-84 is a Rab1 binding partner involved in Golgi structure. Traffic. 2003;4:153–161. doi: 10.1034/j.1600-0854.2003.00103.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Emr S, et al. Journeys through the Golgi - taking stock in a new era. J. Cell Biol. 2009;187:449–453. doi: 10.1083/jcb.200909011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gillingham AK, Tong AHY, Boone C, Munro S. The GTPase Arf1p and the ER to Golgi cargo receptor Erv14p cooperate to recruit the golgin Rud3p to the cis-Golgi. J. Cell Biol. 2004;167:281–292. doi: 10.1083/jcb.200407088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luke MR, Kjer-Nielsen L, Brown DL, Stow JL, Gleeson PA. GRIP domain-mediated targeting of two new coiled-coil proteins, GCC88 and GCC185, to subcompartments of the trans-Golgi network. J. Biol. Chem. 2003;278:4216–4226. doi: 10.1074/jbc.M210387200. [DOI] [PubMed] [Google Scholar]
- 39.Linstedt AD, Hauri HP. Giantin, a novel conserved Golgi membrane protein containing a cytoplasmic domain of at least 350 kDa. Mol. Biol. Cell. 1993;4:679–693. doi: 10.1091/mbc.4.7.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mitoma J, Ito A. Mitochondrial targeting signal of rat liver monoamine oxidase B is located at its carboxy terminus. J. Biochem. 1992;111:20–24. doi: 10.1093/oxfordjournals.jbchem.a123712. [DOI] [PubMed] [Google Scholar]
- 41.Silvius JR, Bhagatji P, Leventis R, Terrone D. K-ras4B and prenylated proteins lacking “second signals” associate dynamically with cellular membranes. Mol. Biol. Cell. 2006;17:192–202. doi: 10.1091/mbc.E05-05-0408. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.