Figure 5.
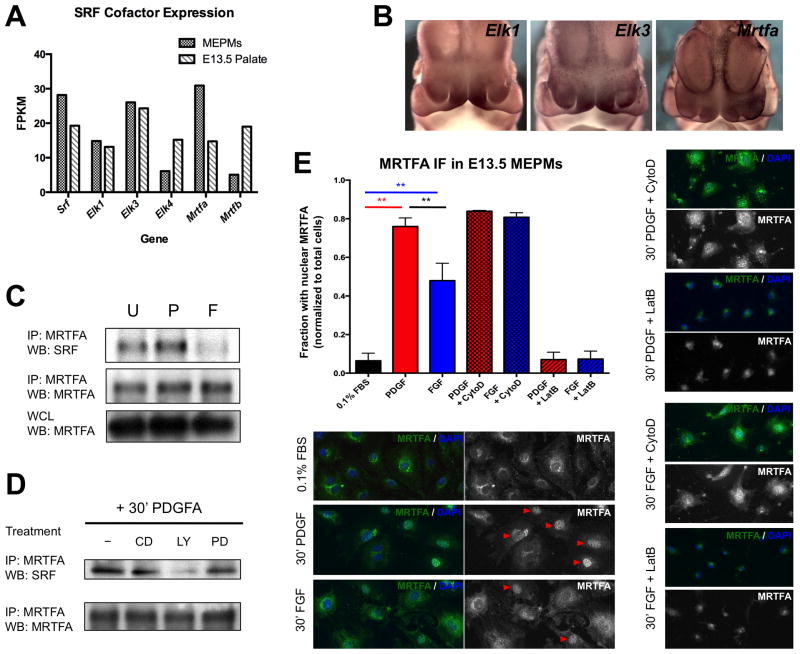
Both MRTF and TCF cofactors play roles downstream of RTK signaling in craniofacial development. (A) Of the five major TCF and MRTF cofactor family members, only Elk1, Elk3, and Mrtfa are expressed above a threshold of 10 fragments per kilobase of transcript per million mapped reads (FPKM) in both E13.5 MEPMs and E13.5 palate. (B) Whole mount in situ hybridization reveals Elk1 and Elk3 mRNA are enriched in the E11.5 medial nasal process (MNP). Mrtfa mRNA expression in the midface is more diffuse but shares expression domains with Pdgfra and Srf. (C) PDGF modestly increases SRF-MRTFA association while FGF reduces SRF-MRTFA complex formation. MRTFA levels are not modulated by PDGF or FGF treatment, thus serving as an additional loading control. (D) MRTFA-SRF association following 30 minutes PDGF stimulation requires PI3K activity. All biochemistry performed in E13.5 MEPMs. CD = cytochalasin D, PD = PD325901, LY = LY294002. U = untreated cells, P = 30 minutes 30 ng/mL PDGFAA, F = 30 minutes 50 ng/mL FGF1 + 1 μg/mL heparin. (E) MRTFA immunofluorescence shows greater nuclear accumulation of MRTFA in response to PDGF compared to FGF, although a significant number of FGF treated cells contain nuclear MRTFA. Red arrowheads mark cells counted as containing nuclear MRTFA. Cytochalasin D and Latrunculin B were used as a positive and negative control, respectively. ** p < 0.05. Data plotted as mean ± SEM. See also Fig. S4.