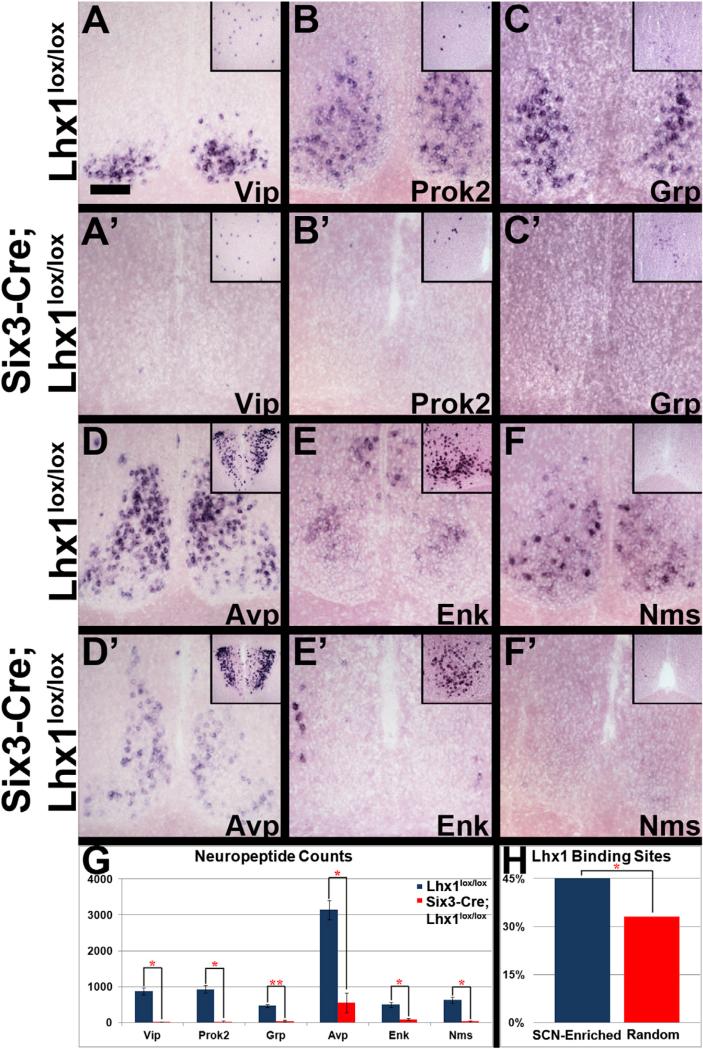
Figure 1. Lhx1 Effects on SCN Neuropeptides.
(A–F’) ISH of SCN from adult Lhx1lox/lox control (A–F) and Six3-Cre;Lhx1lox/lox mutant (A’–F’) mice for Vip (A, A’), Prok2 (B, B’), Grp (C, C’), Avp (D, D’), Enk (E, E’), and Nms (F, F’). Insets show the neocortex (A and C), preoptic area (B, E, and F), and paraventricular nucleus (D). Scale bar represents 100 μm.
(G) Mutants had fewer neuropeptide ISH-labeled SCN cells than controls (n = 3, paired two-tailed t tests, *p < 0.05, **p < 0.01; graph depicts mean ± SEM).
(H) MOPAT bioinformatics analysis showed more putative Lhx1-binding sites in enhancers of an SCN-enriched gene pool compared to a randomly selected pool (pool shown in Table S2; method described in Hu et al., 2008; binomial distribution, *p < 0.05).