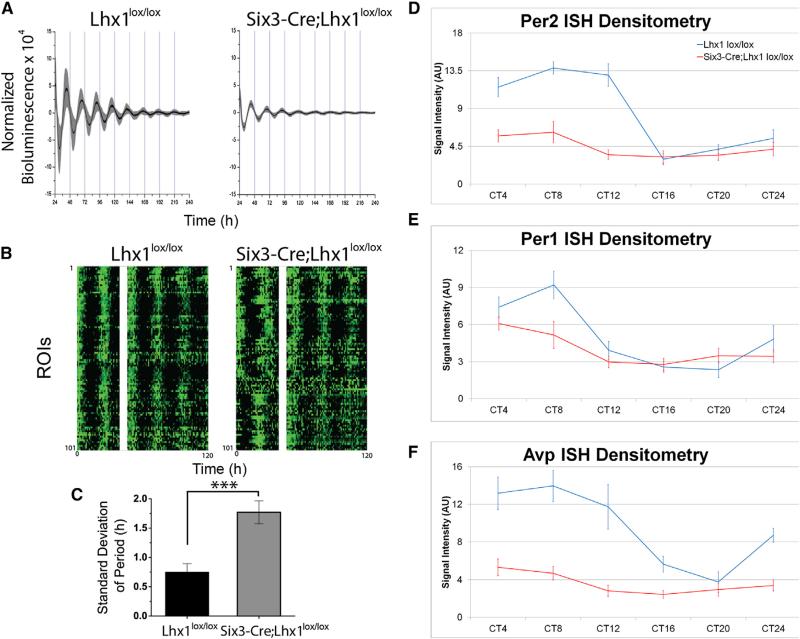
Figure 3. Lhx1 Effects on SCN Clock Gene Rhythms.
(A) Adult Lhx1lox/lox (left) and Six3-Cre;Lhx1lox/lox (right) mouse SCN explants showed near-24 hr cycling of Per2::Luc bioluminescence over 10 days of recording, with moderately damped amplitude in the mutant explants. Traces show mean (black line) and SD (gray) of bioluminescence collected at 1 min intervals from 8 Lhx1lox/lox (left) and 9 Six3-Cre; Lhx1lox/lox (right) explants.
(B) Raster plots demonstrate variations in synchrony of circadian rhythms across regions of interest (ROIs) from one representative control (left) and one Six3Cre;Lhx1lox/lox SCN explant (right). White bars cover the region where data were lost while recording (40–46 hr).
(C) Loss of Lhx1 caused greater variance of circadian period in Six3-Cre;Lhx1lox/lox SCN explants (n = 5) compared to control littermates (n = 8).
(D–F) Circadian variation in expression levels of SCN clock genes Per2 (D), Per2 (E), Bmal1 (C), and Avp (F). ISH labeling intensity was present in adult Lhx1lox/lox control (D–F) and Six3-Cre;Lhx1lox/lox mutant (D’–F’) mice, with genotype and interaction effects for some genes (n = 3, ANOVA).