Figure 2.
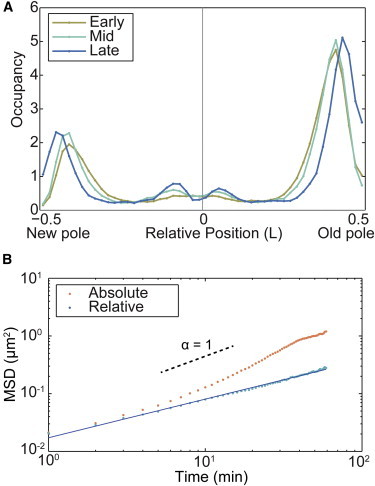
(A) Normalized histogram of the position of MS2-mRNA complexes in relative length units (N = 282,860). Because of cell-to-cell variation in doubling time, cell cycles were partitioned into thirds: early, mid, and late. 76% of the molecular complexes are localized near the poles of the cell. (B) MSD analysis of MS2-mRNA motion. The MSD is computed using two velocity models: absolute (orange) and relative (blue). The relative model MSD has a constant slope with a scaling parameter α = 0.66, consistent with previous measurements, whereas the absolute model increases in slope because of cell growth at times greater than 10 min (N = 8053 trajectories). To see this figure in color, go online.
