Figure 4. Anti-motifs drive negative selection for optimizing specificity.
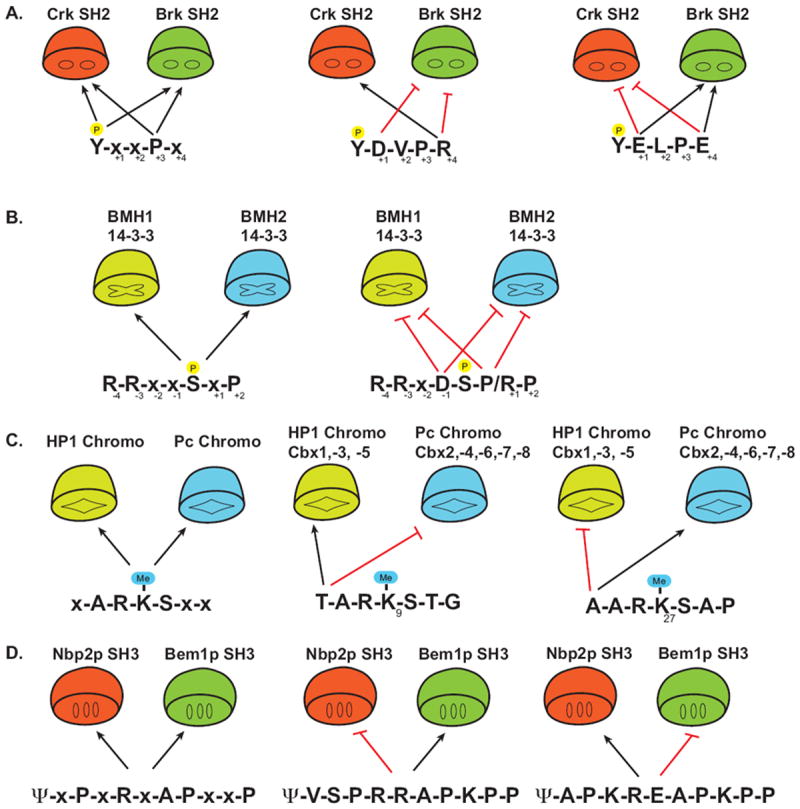
(A) The SH2 domain of Crk and Brk both recognize the motif pY-x-x-L/P (the P in yellow circle above the Y represents a phosphate). Black arrows indicate permissive or positive preferential residues. Anti-motifs (red hash) of select peptides determine the selectivity for either Crk or Brk. (x denotes any natural amino acid) (B) Yeast 14-3-3 proteins BMH1 and BMH2 recognize the consensus motif R-R-x-x-pS-x-P (left panel), however sequences which contain an Asp at -1 or Pro/Lys at +1 disrupt binding despite having permissive factors.
(C) Recognition of the methylated lysines (Me-K) on the histone tail by distinct Chromo domains is determine by anti-motifs. Me-K(9) and Me-K(27) both contain the core consensus x-A-R-K-S-x-x. Recognition by either the HP-1 subclass or the Pc subclass of Chromo domains is determined by the amino acid at the -3 position of the Me-K. (D) The yeast SH3 domains of Bem1p and Nbp2p recognize the consensus motif ψ-x-P-x-R-x-A-P-x-x-P (ψ represents a hydrophobic residue, x is any natural amino acid). The sensing of the sequence context by these SH3 domains either inhibits (red hash line) or allows binding (black arrows), thereby preventing non-specific interactions.
