Figure 1.
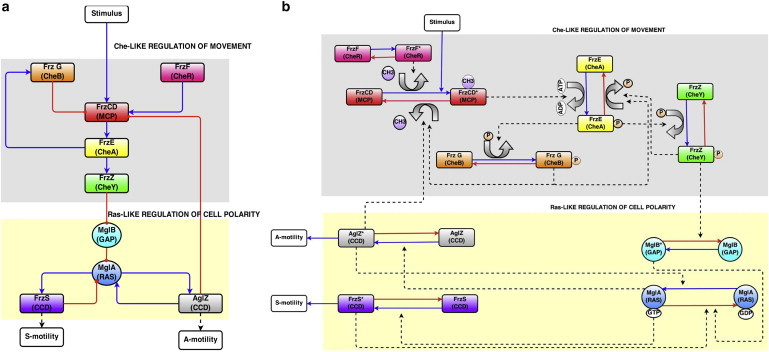
Oscillatory model for regulation of motility in M. xanthus. M. xanthus Frz and Mgl proteins can be depicted as either (a) as a simple circuit diagram or (b) a network diagram of biochemical interactions. The pathway requires the Che-like Frz proteins, and the Ras-like GTPase Mgl for regulating motility. Unlike E. coli, there are no random reorienting tumbles; in M. xanthus the entire cell polarity switches, causing the cell to retread its path. Our model is based on the following steps: An external stimulus (not required) causes a conformational change in the FrzCD receptor that stimulates autokinase activity of FrzE. Phosphotransfer from FrzE∼P to FrzZ causes a dissociation of FrzZ∼P from FrzE. FrzZ∼P disrupts the MglB-MglA complex at the leading pole releasing MglA. A new polarity is established by MglA, which interacts with the coiled coil domain proteins AglZ and FrzS to regulate motor activity of the two motility systems at the leading pole. FrzF methyltransferase activity and feedback from FrzG onto FrzCD adapt the system. Feedback from AglZ to FrzCD, and from the motors on to MglA form a dual oscillation cycle. To see this figure in color, go online.
