Figure 3.
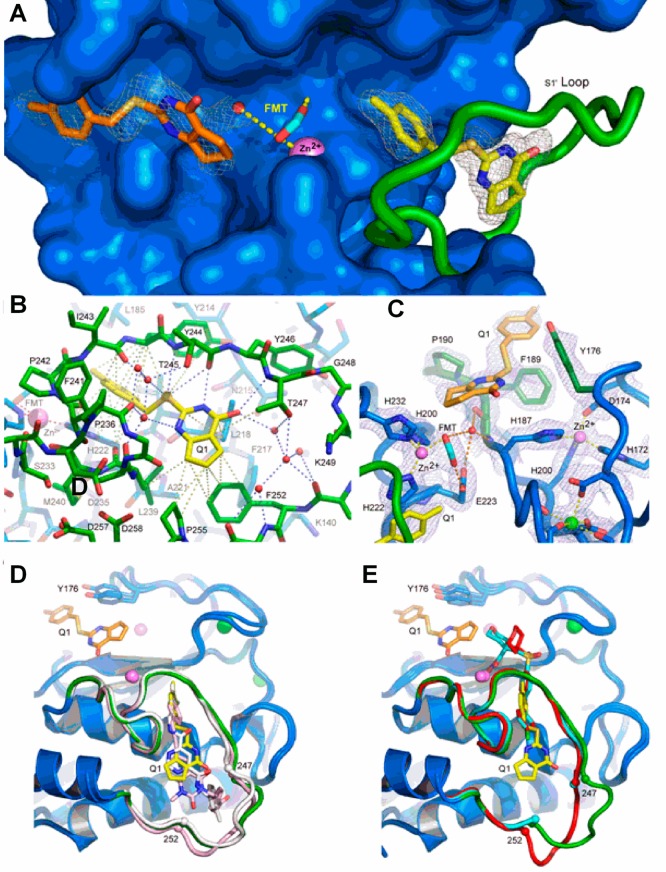
Structure of the MMP-13 CAT–compound 2 complex revealing the distinct binding sites in protomer A (see the text). (A) Annealed omit map with coefficients Fo – Fc contoured at 2.5σ superimposed on the refined model of the MMP-13 CAT–2 complex. 2 was left out of the phase calculation. The surface of the protein is blue, and the S1′ loop is green. The catalytic zinc ion is a violet sphere, and formate from the reservoir solution is shown as cyan sticks. The two distinct 2 molecules are represented as yellow and orange sticks. Formate from the reservoir solution chelates the zinc ion and forms hydrogen bonds with a nearby water molecule and the side chain of the catalytic glutamic acid (E233) hidden beneath the surface. (B) S1′ 2 binding site in MMP-13 CAT. The color scheme is the same as that in panel A, and the view is rotated only slightly around the vertical compared to that of panel A. Gray dashed lines represent selected van der Waals contacts (<4.6 Å), and the blue dashed lines represent hydrogen-bonding interactions. (C) σ–A weighted electron density with coefficients 2mFo – dFc superimposed on the S1/S2* 2 binding site in the refined model of the complex. The color scheme is the same as that in panel A except Y176, F189, and P190, which make the majority of contacts with 2, are highlighted as dark green sticks. The green sphere is a calcium ion. Hydrogen bonds are shown as orange dashes, and metal–ligand interactions are shown as yellow dashes. (D) Superposition of compounds 2 (yellow), 4 (white), and 5 (pink) in the S1′ binding site. All three compounds accept a hydrogen bond from the amide nitrogen of Thr247. A second molecule of 2 can be seen on the other side of catalytic zinc in the S1/S2* site. (E) Superposition of compound 2 (yellow) with hydroxamic acid-based inhibitors (PDB code 456C, red; PDB code 830C, cyan36) that do not intrude deeply into the S1′ specificity loop. The difference in positions of residues 248–251 (disordered in structure 830C) in the two classes of inhibitors suggests the S1′ specificity loop is conformationally dynamic in the uninhibited enzyme.