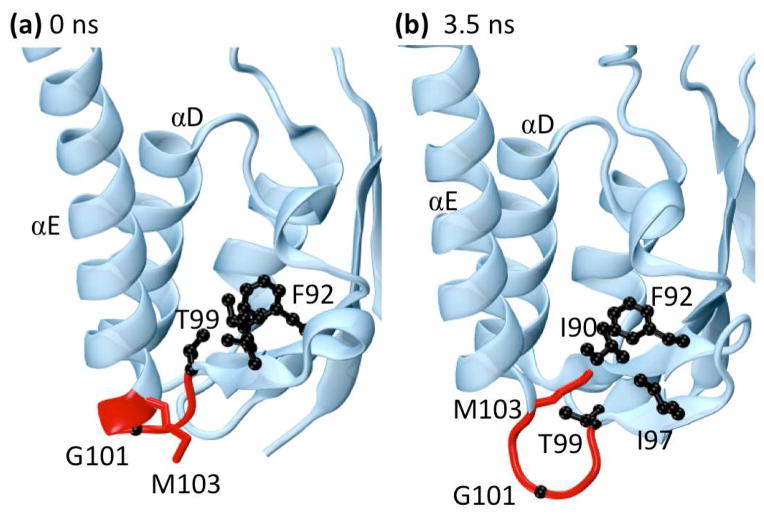
Fig. 7.
The protein γδ resolvase (PDB: 1gdt). The side-chains of residues forming a hydrophobic pocket (I90, F92, I97, T99, and M103) are shown in black while the backbones of residues 99–103 are shown in red. (a) Residues near loop 5E in the minimized crystal structure. (b) Residues near loop 5E at 3.5 ns. Near 3.5 ns the end of helix αE and part of loop 5E unwind to form an Ω-loop. This motion flips the side-chain of residue M103 into the hydrophobic pocket shown in black while pushing residue G101 into solvent, stabilizing the alternate conformation. Both M103 and G101 are known to be important for the binding and flexibility of αE and were identified as highly significant during this time range by wavelet analysis.