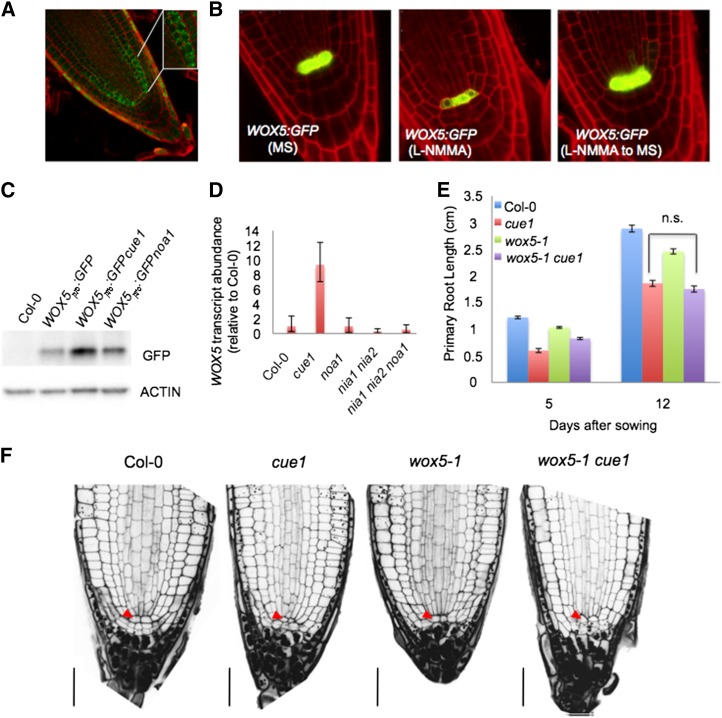
Figure 6.
Genetic analysis between NO altered mutant backgrounds and wox5. A, Detection of endogenous NO production using DAF-2DA in 5-d-old wild-type (Col-0) roots. The RNS-dependent biosensor fluoresces strongly in cortex/endodermis initials (inset) and the immediate progeny generating endodermis and cortex tissues in meristem cells. B, WOX5 marker analysis in wild-type (Col-0) roots grown in the absence or presence of 20 µm L-NMMA and then transferred to MS medium. C, Immunoblot analysis with anti-GFP antiserum of in vivo levels of GFP protein in root extracts of WOX5pro:GFP, WOX5pro:GFPcue1, and WOX5pro:GFPnoa1 5-d-old seedlings. Actin protein levels also were determined as a loading control. D, WOX5 transcription levels in roots of the wild type (Col-0) and cue1, noa1, nia1 nia2, and nia1 nia2 noa1 mutants. E, Primary root lengths in the 5- and 12-d-old wild types (Col-0) and 5- and 12-d-old cue1, wox5-1, and wox5-1 cue1 mutants. Error bars represent se; n > 40. n.s., No statistically significant difference. F, Root sections of 5-d-old wild-type (Col-0), cue1, wox5-1, and wox5-1 cue1 seedlings. Red arrowheads indicate QC. Bars = 30 µm.