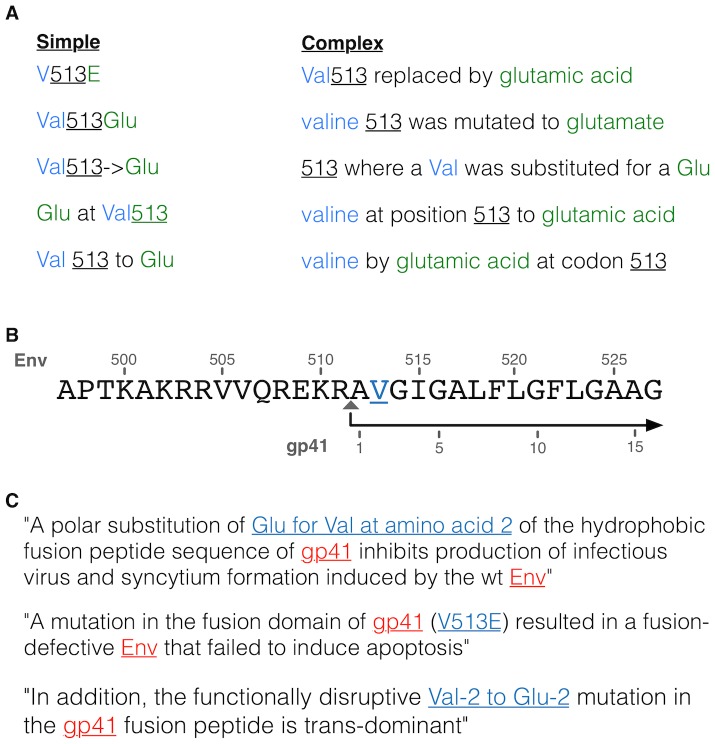
Figure 4. Example illustrations of issues associated with parsing and mapping mutation data.
(A) Representative simple and complex examples of sentences recognised by the templates used to perform the mutation text-mining of articles (see Table S1 for complete list). Information of interest representing the wildtype residue (blue) and mutated residue (green) are coloured and position of the mutations are underlined. (B) Illustration of the distinct numbering schemes for different chains of the same protein. The shown peptide sequence is a short region (497–527) of the HIV Envelope glycoprotein gp160 (Env) overlapping the site cleaved by the host furin to produce the Surface protein gp120 and Transmembrane protein gp41 chains. The cleavage site is denoted by a grey triangle. The numbering above the sequence defines the position relative to the start of the gp160 protein and the numbering below the sequence defines the position relative to the start of the gp41 chain. (C) Examples of three sentences from the HIV literature where each article uses a different nomenclature or numbering scheme (blue) to describe the same mutation at the same site, Valine at residue 513 in the gp160 protein. Each article also refers to the protein by the chain name, gp41, rather than the name of the unprocessed protein, Envelope glycoprotein gp160 (Env), used for mapping in the HIV mutation resource. One example sentence refers to the gp41 chain while utilising the numbering for the unprocessed protein.