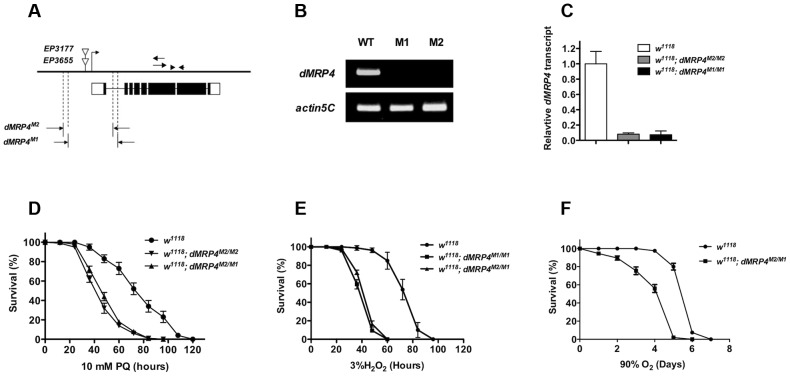
Figure 2. dMRP4 is required for oxidative stress resistance.
(A) Molecular analysis of dMRP4 mutants. The solid bar represents the genomic region of dMRP4. The bent arrow indicates the transcription start site of dMRP4 gene. The open triangles show the insertion positions of EP3655 and EP3177. Open boxes below the solid bar represent exons of dMRP4 transcript and filled boxes indicate the encoding protein sequences. The span of both deletions was determined by sequencing the corresponding regions with specific primers. The arrows were primers for dMRP4-related semi-quantitative RT-PCR and arrowheads for qt-PCR experiments. The deleted sequences were described in Materials and Methods . (B) Expression of dMRP4 mRNA in two mutant alleles. Semi-quantitative RT-PCR was used to determine the levels of dMRP4 mRNA expression. dMRP4 mRNA was under-detectable in dMRPM1/M1 (M1) or dMRPM2/M2 (M2). Actin5C served as an internal standard. (C) qt-PCR analysis of dMRP4 mRNA in two dMRP4 alleles. (D) Effects of paraquat-induced oxidative stress on dMRP4 mutant flies (n = 180 for each group). (E) Effects of hydrogen peroxide-induced oxidative stress on dMRP4 mutant flies (n = 200 for each group). (F) Effects of hyperoxia-induced oxidative stress on dMRP4 mutant flies (n = 200 for each group). Error bars represent S.E.