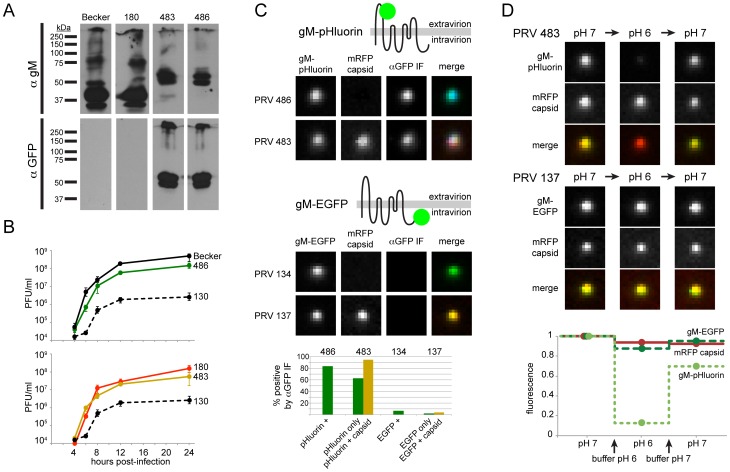
Figure 1. Characterization of PRV recombinants.
(A) Cells infected with parental viruses (PRV Becker, 180) or gM-pHluorin expressing viruses (PRV 483, 486) were harvested at 12 hpi, and lysates were analyzed by Western blot. Parallel blots were probed with polyclonal antiserum against gM (αgM), or a monoclonal antibody that recognizes pHluorin (αGFP). (B) Single-Step Virus Replication. Parallel cell cultures were infected in triplicate with the indicated parental viruses (PRV Becker, 180), gM-pHluorin expressing viruses (PRV 483, 486), or a gM-null virus (PRV 130). Cells and supernatants were harvested at indicated times, and infectious virus titer was measured by plaque assay. Error bars represent range. (C) Membrane Topology. Particles produced by the indicated viruses were imaged to detect gM-pHluorin or gM-EGFP, mRFP capsid, and immunofluorescence targeting the pHluorin or EGFP epitopes (αGFP IF). Immunofluorescence labeling was performed without membrane permeabilization. The schematic represents the predicted topology of gM-pHluorin or gM-EGFP. Images depict single representative virus particles (each image is 2.5 µm by 2.5 µm). Bar graph represents classification and quantification of particles based on fluorescence (n≥237 particles per condition). (D) pH Sensitivity. Particles produced by the indicated viruses were imaged to detect gM-pHluorin or gM-EGFP, and mRFP capsid after addition of buffers at pH 6 or 7. Images depict single representative virus particles (each image is 2.5 µm by 2.5 µm). Graph represents relative particle fluorescence after each indicated buffer change (n≥154 particles per condition).