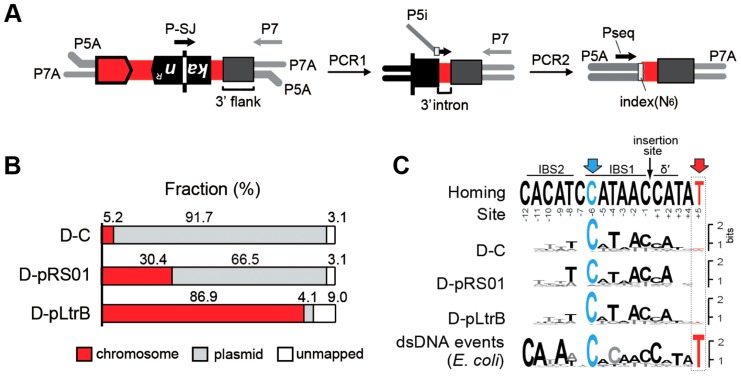
Figure 3. Mapping of intron insertion sites with RIG-seq.
(A) RIG-seq amplification scheme. High-throughput targeted sequencing of insertion loci was based on the generation of splice junction (SJ) sequence of the kanR gene during retrotransposition. After ligation of P5A and P7A, Illumina-specific adapters, two tandem PCRs were used to amplify 3′ flanks of retrotransposition events in Illumina libraries: a specific SJ primer (P-SJ) with the P7 primer in the first PCR; then library-specific primers (P5i) carrying library-specific indices with the P7 primer in the second PCR. Pseq, sequencing primer. (B) Replicon preferences. Mapping of the Illumina libraries demonstrated a marked preference for intron insertion into the donor plasmid pLNRK-RIG in control D-C strain (Figure S2 in Text S1); the overwhelming majority of the reads for D-pLtrB was mapped to the chromosome. (C) Insertion site sequence logos compared to the homing-site sequence. The logos were generated based on multiple sequence alignments of retrotransposition events. The homing site sequence with intron-binding sites (IBS1 and IBS2) and Δ′, required for reverse splicing, is shown on the top. The most conserved residue C-6 is highlighted in blue. The T+5 is shown in red and represents the nucleotide indicative of endonuclease-dependent events. The low frequency of T+5 suggests that endonuclease-independent pathways into ssDNA dominate in L. lactis IL1403. The logo for endonuclease-dependent dsDNA events identified in E. coli is shown for comparison at the bottom [24].