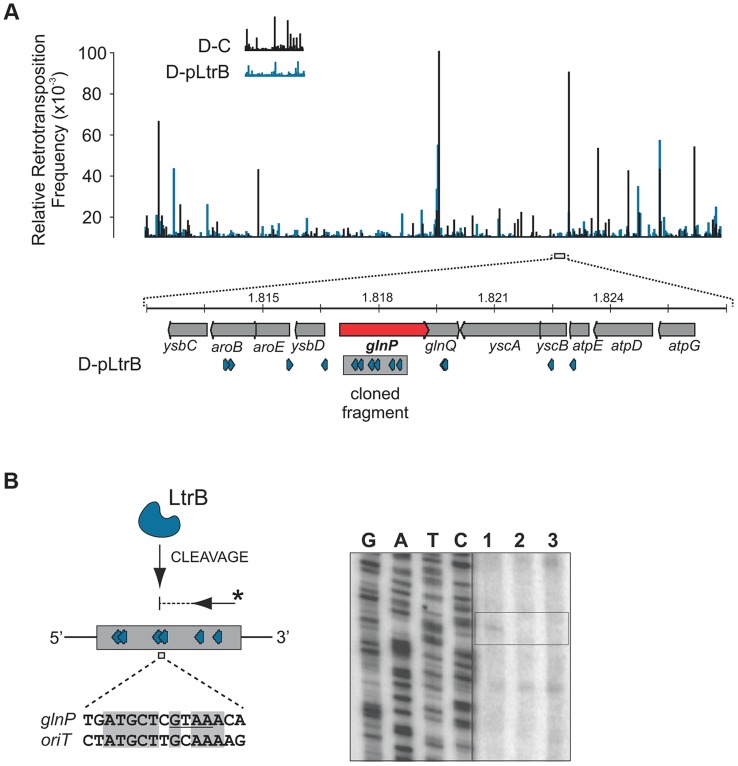
Figure 6. LtrB has off-target activity.
(A). Mapping of sites and selection of the target sequence. Randomly sampled libraries of the reads for D-C and D-pLtrB were mapped along the L. lactis IL1403 chromosome [25] (Tables S2 and S4 in Text S1). The relative retrotransposition frequencies are represented by vertical bars: black, D-C strain without relaxase; blue, pLtrB strains with relaxase. For detection of possible relaxase off-target nicking activity, we chose the glnP gene (red arrow), in which six different retrotransposition events were observed in the D-pLtrB strain (blue), but none in control strain D-C. (B) LtrB relaxase has weak off-target activity. The ssDNA containing a fragment of the glnP gene was incubated with either LtrB-HIS6 (lane 1), catalytic mutant LtrB (Y21A)-HIS6 (lane 2) or left untreated (lane 3). Primer extension reactions (shown schematically) were used to detect cleavage sites. Weak relaxase-specific bands were detected (see also S4 Figure in Text S1). One of the cleavages was mapped between sites for two retrotransposition events detected in the D-pLtrB library. The cleaved sequence is similar to the oriT nicking site of pRS01 as shown by pairwise sequence alignment (bottom).