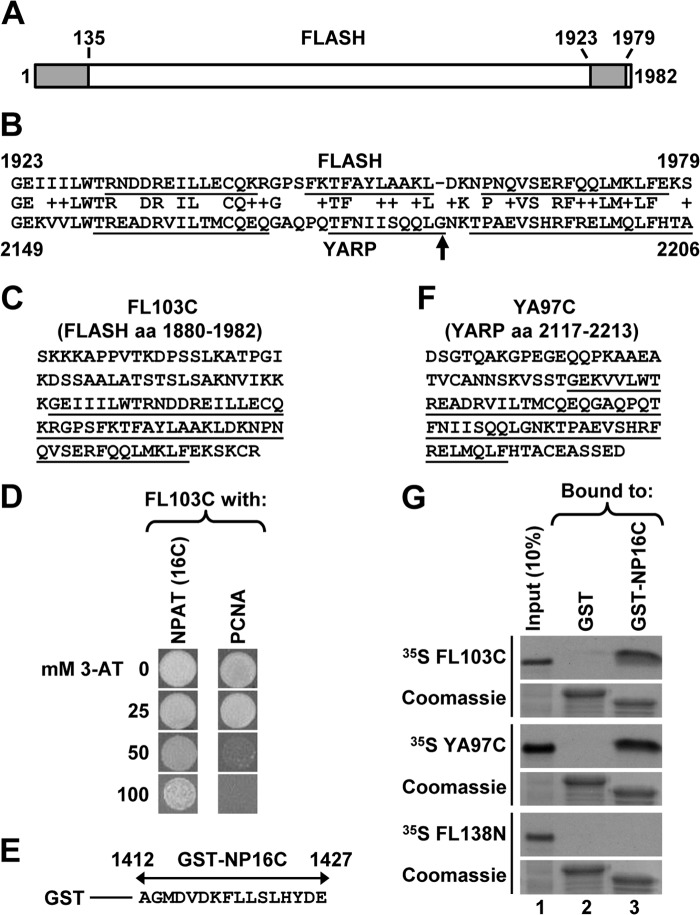
FIGURE 1.
C-terminal regions of FLASH and YARP interact with NPAT. A, diagram of human FLASH consisting of 1982 amino acids. Positions of the conserved N- and C-terminal domains are indicated with gray boxes. B, protein BLAST alignment of the C-terminal domains of FLASH (top, amino acids 1923–1979) and YARP (bottom, amino acids 2149–2206). Amino acid substitutions that are unlikely to change the overall protein structure, as determined by the BLOSUM62 scoring matrix, are marked by “+”. α-Helices, as predicted by NMR structural studies (see “Discussion” for details), are underlined. The highly conserved glycine present in vertebrate YARP is indicated with an arrow. C, sequence of the last 103 amino acids of human FLASH. The C-terminal domain shared with YARP is underlined. D, yeast two-hybrid screen identifies NPAT and PCNA as potential binding partners of the FLASH C-terminal region. Growth of yeast cells expressing the C-terminal region of FLASH and either the last 16 amino acids of NPAT (left vertical row) or the C-terminal half of PCNA (right vertical row) in the presence of increasing concentration of 3-AT. E, sequence of the last 16 amino acids of human NPAT fused to the N-terminal GST (GST-NP16C). F, sequence of the last 97 amino acids of human YARP. The C-terminal domain shared with FLASH is underlined. G, GST pulldown assay to analyze in vitro interaction of 35S-labeled C-terminal regions of FLASH (FL103C) or YARP (YA97C) with the last 16 amino acids of NPAT fused to GST (GST-NP16C). 35S-Labeled N-terminal region of FLASH (FL138N, amino acids 1–138) and GST alone were used as negative controls. The amount of each GST protein purified on glutathione beads was monitored by staining the gel with Coomassie Blue (bottom panels). Note that GST alone migrates higher than GST-NP16C due to containing random amino acids at the C terminus from translating the multiple cloning site present in the pET-42a vector.