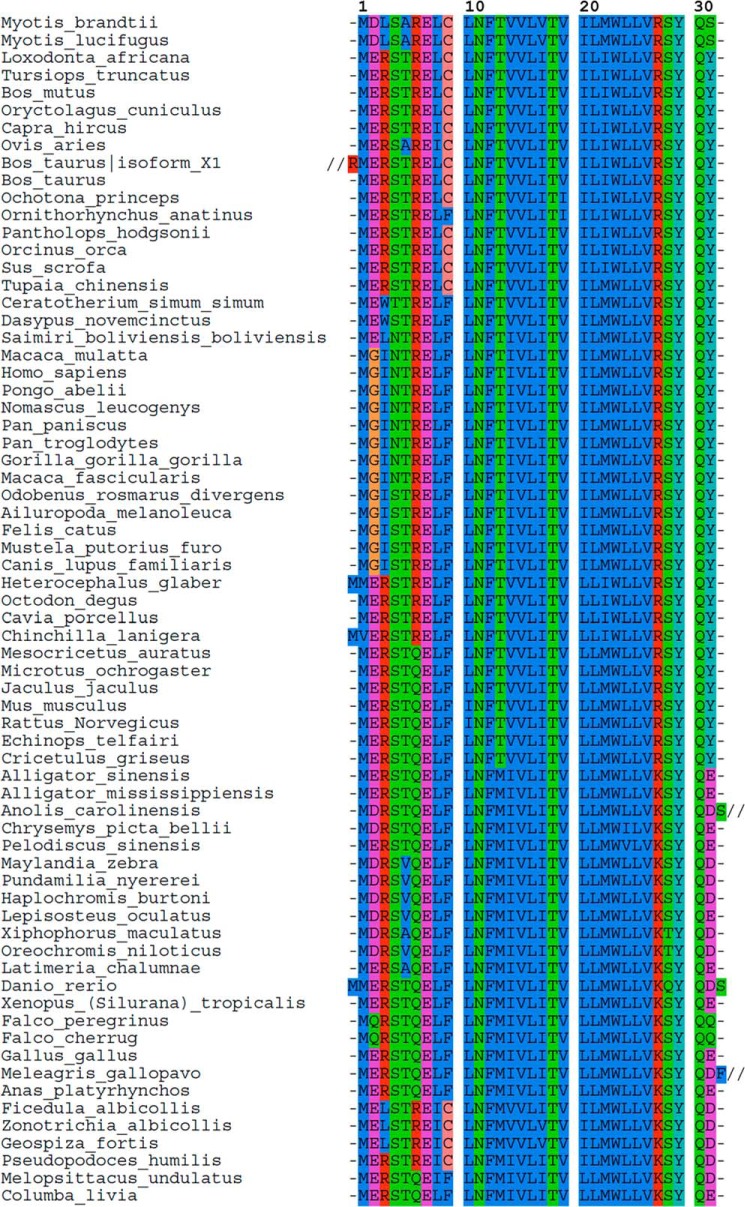
FIGURE 7.
Alignment of available sarcolipin peptide sequences. The human SLN sequence (Uniprot no. O00631) was used as a template for a BLASTp search in the non-redundant protein sequence database (55). The PAM-30 algorithm was used as it is recommended for analysis of short peptide sequences. Following this strategy, 110 sequences were retrieved from the resulting taxonomy report, covering 67 species. However, when several sequences were available in the database for a given species (e.g. 13 sequences for human), all the available sequences were exactly the same, except for the cow and the ferret. In these two cases, a second SLN isoform was predicted from whole genome sequencing. Those sequences are identical in the membrane part but have putative additional residues in the N and/or C termini. The alignment presented here was done with the Seaview program (56, 57). Three sequences were shortened to optimize the figure presented here: Bos taurus isoform_X1, residues M1SSLFASQSEKALPRAQKSRQPGRQEGVGAQVGEREEKPIAAQCVLDPIQEVRTSRRSLVCPQKFAFSSHQATL75; Anolis carolinensis, residues R33KEARRCGEWGAGFLMPPMMQGNLVAE59; Meleagris gallopavo, residues L33FAKDTLEERWLSVFTGFCHQPFEV57.