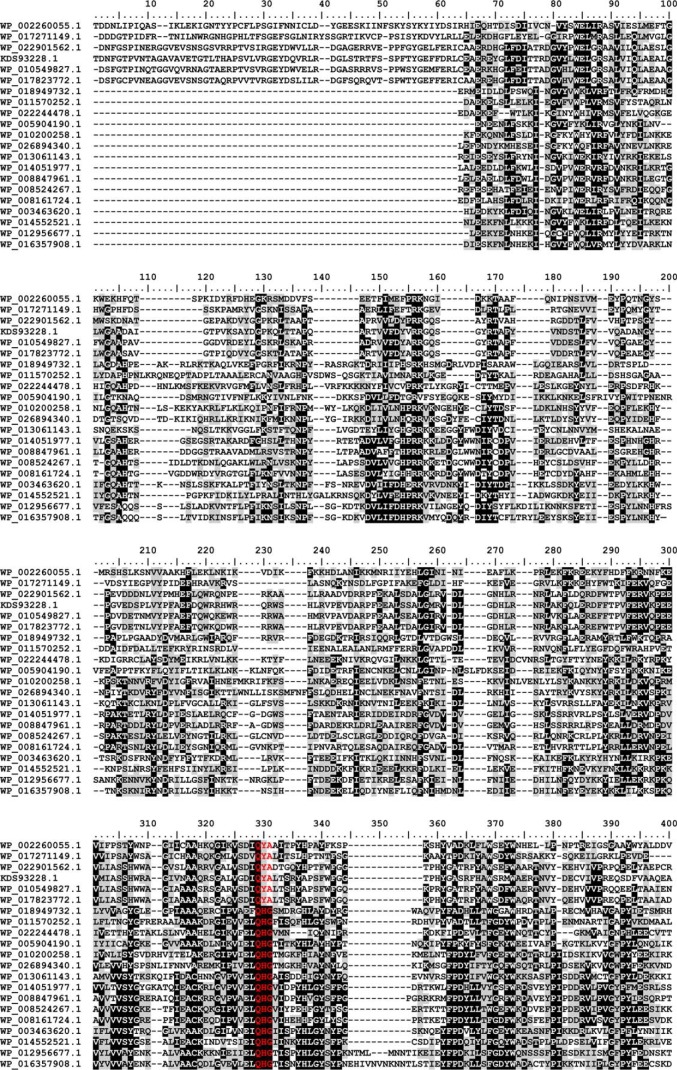
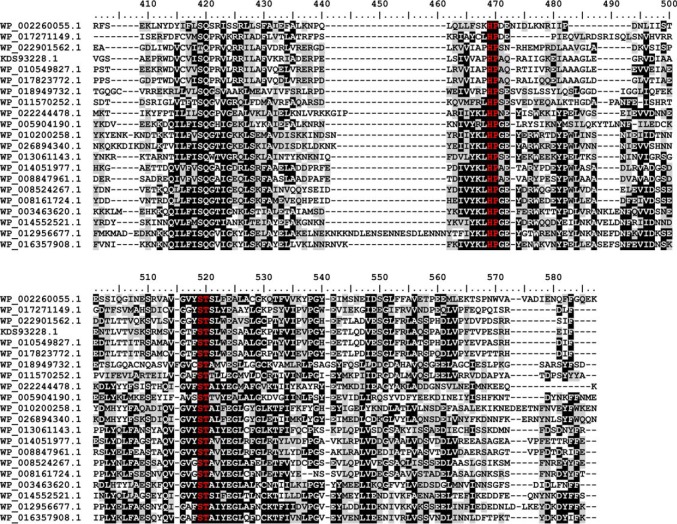
FIGURE 7.
Multiple sequence alignment of family GT97. Protein sequences were aligned with the MAFFT server (G-INS-I strategy; BLOSUM20 scoring matrix; unalignlevel: 0.4) (21) and visualized using BioEdit (42). High conservation is indicated by white letters on a black background, less conserved regions have black letters on a gray background, and the sialyl-motifs are highlighted in red. Accession numbers correspond to SiaDW and uncharacterized proteins from the following organisms: WP_002260055.1 (aa 563–1037/N. meningitidis serogroup W), WP_017271149.1 (aa 545–999/Sinorhizobium meliloti), WP_022901562.1 (aa 653–1117/Humibacter albus), KDS93228.1 (aa 678–1144/Dermabacter hominis 1368), WP_010549827.1 (aa 651–1116/Brachybacterium paraconglomeratum), WP_017823772.1 (aa 664–1129/Brachybacterium muris), WP_018949732.1 (aa 18–458/Thioalkalivibrio sp. ALMg11), WP_011570252.1 (aa 101–539/Roseobacter denitrificans), WP_022244478.1 (aa 8–439/Roseburia sp. CAG:45), WP_005904190.1 (aa 21–464/Fusobacterium nucleatum), WP_010200258.1 (aa 9–467/Bacillus sp. m3-13), WP_026894340.1 (aa 8–467/Clostridiisalibacter paucivorans), WP_013061143.1 (aa 8–446/S. ruber), WP_014051977.1 (aa 44–483/halophilic archaeon DL31), WP_008847961.1 (aa 33–474/H. kocurii), WP_008524267.1 (aa 8–446/H. tiamatea), WP_008161724.1 (aa 9–445/N. sulfidifaciens), WP_003463620.1 (aa 15–456/Gracilibacillus halophilus), WP_014552521.1 (aa 15–457/Halanaerobium praevalens), WP_012956677.1 (aa 15–497/Methanobrevibacter ruminantium), WP_016357908.1 (aa 13–470/Methanobrevibacter sp. AbM4).