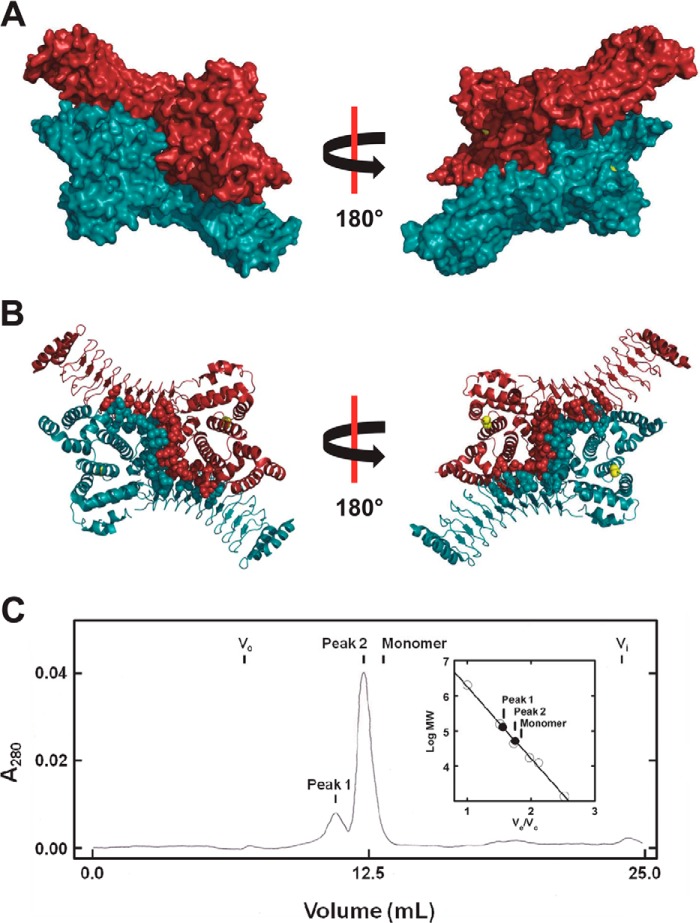
FIGURE 6.
Identification of a symmetric dimer in the crystal structure of the related IpaH3 ligase. A, surface rendering of the crystal structure for IpaH3 (PDB code 3CVR) in complex with its dimer mate identified by PISA analysis. Active site cysteine residues are colored yellow. B, ribbon diagram of the crystal structure for IpaH3 in complex with its dimer mate in the same orientation as in A. Residue side chains predicted by PISA analysis to be at the dimer interface are modeled as spheres. Active site cysteine residues are modeled as yellow spheres. C, gel filtration chromatography of IpaH9.8(244–545), as described under “Materials and Methods.” Inset, relative mobility of peaks 1 and 2 (closed circles) are shown on the calibration plot summarizing a series of standards (open circles). The predicted elution volume for monomeric IpaH9.8(244–545) is indicated (inset).