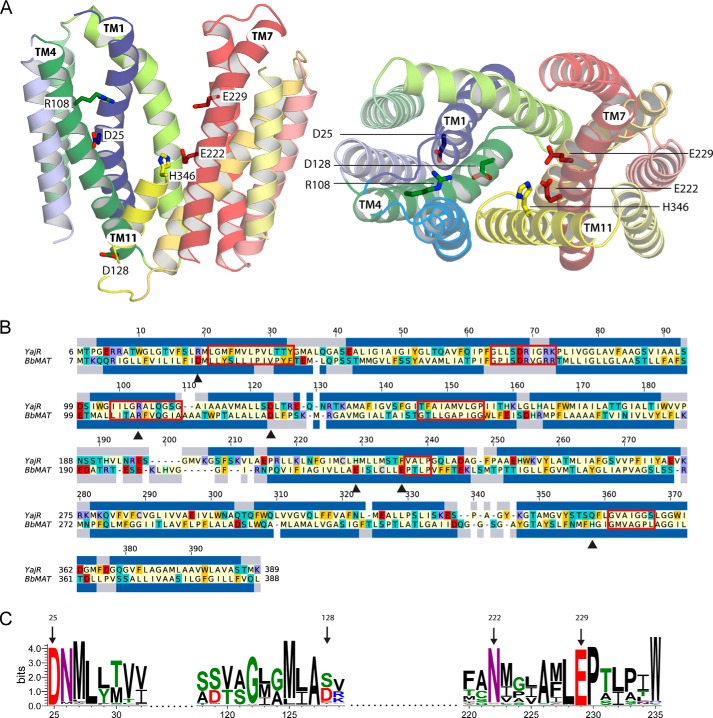
FIGURE 3.
Structural model of BbMAT on the basis of the YajR transporter structure. A, the BbMAT model in an outward-facing conformation is represented as helical cartoons, viewed from the plane of the membrane with the periplasm at the top (left, with TM4 and part of TM11 omitted for clarity) and from the periplasm (right). The model predicts four negatively charged and two positively charged residues exposed to the central cavity, which could be involved in the transport activity of BbMAT. Side chains of these residues are shown as sticks. The figures were generated using PyMOL v1.7.05 (Schrödinger). B, sequence alignment between YajR and BbMAT used for modeling. The alignment was colored according to the chemical nature of the residues as described in Fig. 1. The helices assigned for the YajR structure using DSSP (67) and the PSIPRED prediction for helices in BbMAT are shown as blue bars above and below the sequences, respectively. Red rectangles mark the MFS and DHA12 motifs (10, 25), and black triangles indicate the residues that have been investigated in this study. C, sequence logo illustrating conservation of Asp-25, Asp-128, Glu-222, and Glu-229 among 500 homologues belonging to the MFS family. Multiple sequence alignment was performed using Muscle, and the logo was generated using Weblogo 3.3 (68). The residue colors are as follows: polar (Gly, Ser, Thr, Tyr, and Cys) in green, neutral (Gln and Asn) in purple, basic (Lys, Arg, and His) in blue, acidic (Asp and Glu) in red, and hydrophobic (Ala, Val, Leu, Ile, Pro, Trp, Phe, and Met) in black. Numbering is according to the sequence of BbMAT.