Fig. 4.
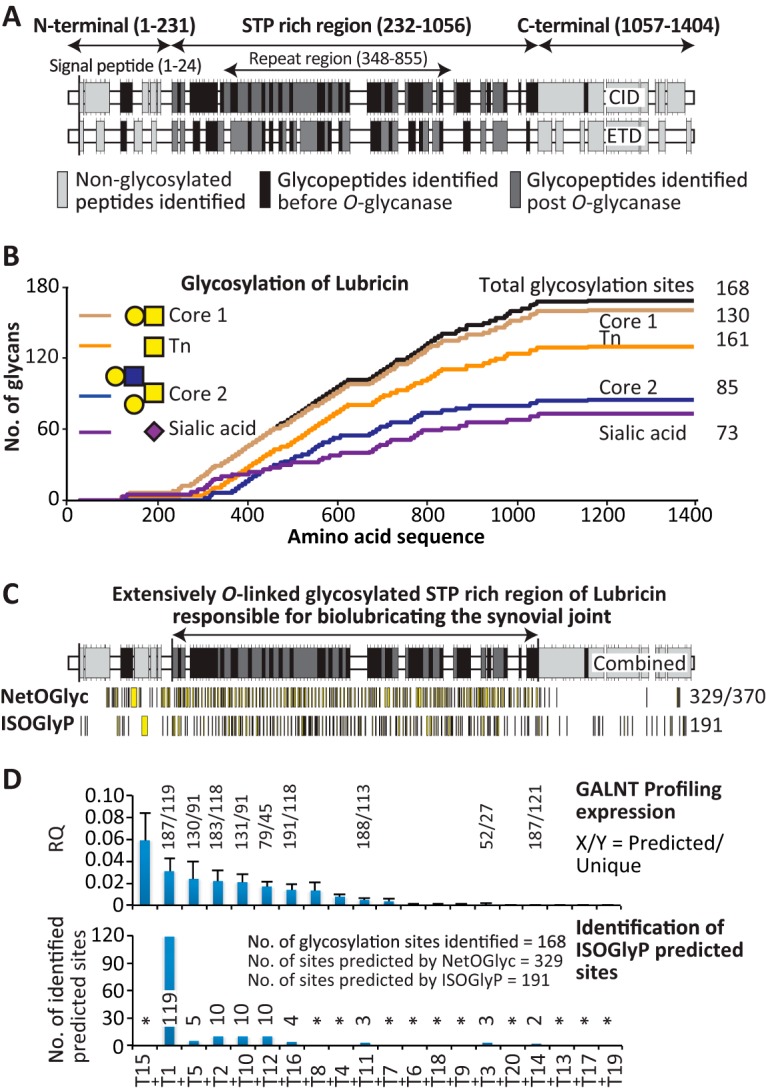
The O-glycosylation map of lubricin. A, the glycosylated peptides identified in the STP-rich region and non-glycosylated peptides (light gray) in the end domains of the lubricin sequence before (black) and after (dark gray) partial de-glycosylation using CID and ETD fragmentation. B, graphic representation of all of the identified glycans and their positions in the protein sequence. The figure includes the identified glycopeptide containing GalNAcα1- (dark orange, 130), core 1 Galβ1–3GalNAcα1- (chocolate, 161), core 2 Galβ1–3(Galβ1–4GlcNAcβ1–6)GalNAcα1- (blue, 85), and sialylated core 1 and 2 (pink, 73) glycans with their positions in the protein sequence. The graph also includes the total number of identified glycosylation sites (168 sites) in the protein sequence (black). The total number of sites is based on the identified peptides and the fact that two threonines in the imperfect tandem repeat variants (e.g. EPAPTTPK, SAPTTPK, EPAPTTTK) can be glycosylated. It indicates a high number of glycosylations in the STP-rich region, whereas the N- and C-terminal regions are scarcely glycosylated. C, the combined CID and ETD identified glycopeptides in the STP-rich region (black and dark gray) and non-glycosylated peptides (light gray) in the end domains of lubricin before (black) and after partial de-glycosylation (dark gray). The software used (NetOGlyc4.0 and ISOGlyP) predicted sites of lubricin O-glycosylated Ser/Thr in the protein sequence (yellow) and the total number of potential Ser/Thr sites (370). The results indicate that the gene-based ISOGlyP software predicted a similar number of glycosylation sites (191 sites) as identified by the data presented in this report (168 sites). D, the GALNT profiling expression analysis of the 20 known human GALNT genes (top) in primary FLSs and the ISOGlyP-predicted sites by the available GALNT genes identified by the data presented in this report (bottom). The genes were arranged in descending order of their expression. RQ, relative quantification. Asterisk denotes genes not included in the ISOGlyP software.
