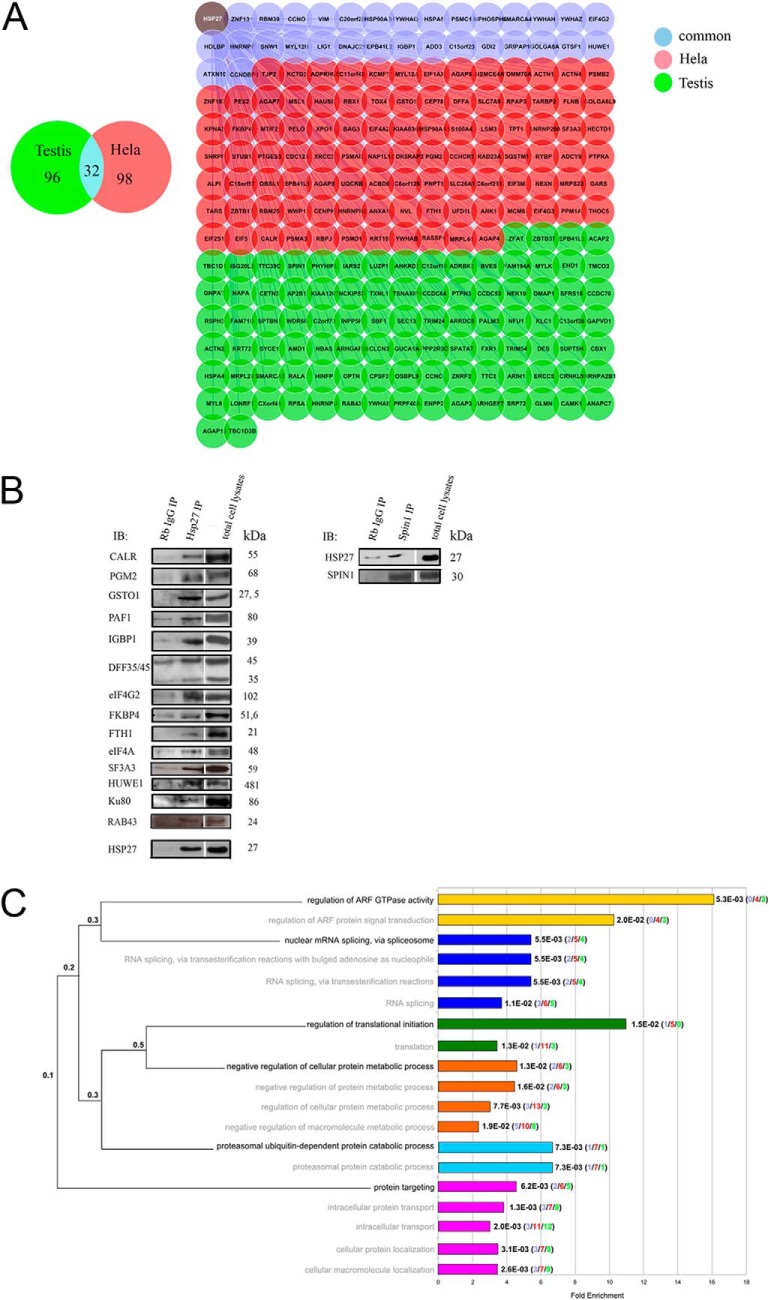
Fig. 1.
Large-scale Y2H screen deciphers Hsp27 PPI. A, The 226 Hsp27-interacting proteins identified in the Y2H screen of HeLa cDNA library (red nodes), testis cDNA library (green nodes), or in common in both libraries (blue nodes). B, Validation of 6% of the Hsp27 interactors by Co-IP from PC-3 cells total protein extracts. Rb (Rabbit), IgG (Immunoglobulin G), IP (Immunoprecipitation), IB (Immuno-blot). C, GO Biological Process enrichments (Benjamini-corrected p values (q-value) < 0.02) of the Hsp27 interaction partners. Barplots on the right indicate the fold enrichment of each annotation, and the exact q-values. Each term corresponds to a colored bar and related terms share the same color. In brackets are represented the numbers of interactors identified from the testis (green), HeLa (red), or both (blue) libraries. The dendrogram on the left depicts the hierarchical organization of the significantly over-represented terms in the ontology. The most precise terms are written in black and the less specialized terms in gray, with the associated term precision at the branching points.