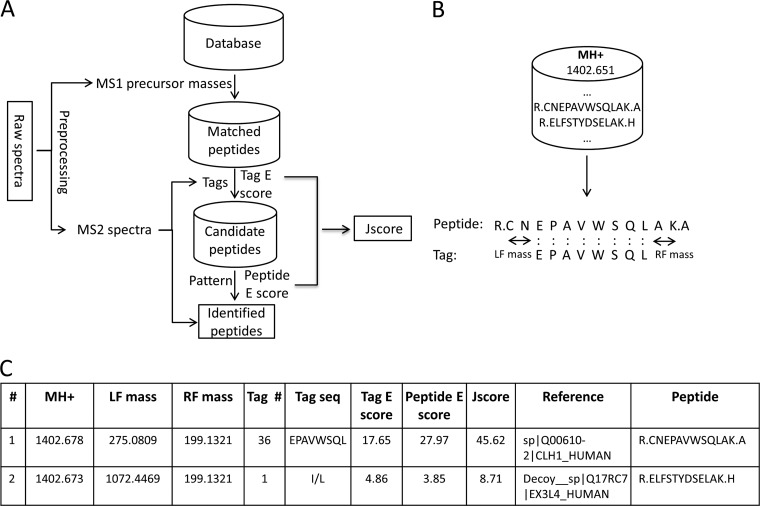
Fig. 1.
The JUMP algorithm. A, overview of the JUMP algorithm. JUMP starts with a raw data file or an mzXML file and then performs MS1 and MS2 preprocessing for each spectrum. Both MS1 precursor ion mass and sequence tags derived from an MS2 spectrum are used to filter peptide sequences in the database. After pattern matching, JUMP generates a final score, the J score, to rank all identified peptides. B, an example to illustrate two-step filtering of theoretical peptides in the JUMP algorithm: (i) filtered by a precursor mass of 1402.651 Da within a mass tolerance range (e.g. 10 ppm); (ii) filtered by a sequence tag. C, an example of JUMP output. The J score is generated by integrating the tag E score and peptide E score.