Figure 4.
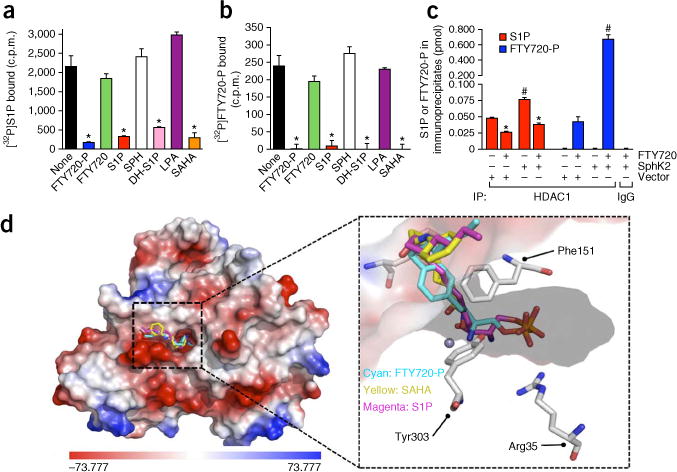
FTY720-P competes with S1P for binding to HDAC1. (a–c) Recombinant His-tagged HDAC1 immobilized to Ni-NTA resin or control Ni-NTA resin (control) was incubated with 0.1 nM [32P]S1P (a) or 0.1 nM [32P]FTY720-P (b) in the absence or presence of unlabeled 1 μM FTY720-P, FTY720, S1P, dihydro-S1P (DH-S1P), sphingosine, LPA or SAHA. Beads were washed extensively and [32P]S1P or [32P]FTY720-P bound to HDAC1 was eluted with 500 mM imidazole and radioactivity detected by scintillation counting. (c) Nuclear extracts from HeLa cells transfected with vector or SphK2 and treated without or with FTY720 (5 μM) for 4 h were immunoprecipitated with anti-HDAC1 antibody or control immunoglobulin G, and bound sphingolipids and FTY720-P determined by LC-ESI-MS/MS. Data are averages of triplicate determinations ±s.d. *P < 0.0001 as compared to none (a,b) or untreated (c) and #P < 0.0001 as compared to vector (unpaired Student’s t-test) (N = 3 independent cell cultures per group). (d) Docking of FTY720-P into the pocket of HDAC2. The low-energy conformations of FTY720-P, S1P and SAHA were calculated when docked in a model based on the crystal structure of HDAC2 (Protein Data Bank: 3MAX). Left, surface contour of the binding site with S1P, SAHA and FTY720-P. The surface contour is colored by electrostatic potential. Right, the binding of S1P (magenta), SAHA (yellow) and FTY720-P (cyan) with HDAC2. The active site residues involved in the interactions with small molecules are shown as sticks and the zinc atom as a sphere.
