Abstract
Background
Epigenome-wide association studies are emerging in the field of cancer epidemiology with the rapid development of large-scale methylation array platforms. Until recently, these methods were only valid for DNA from fresh frozen (FF) tissues. Novel techniques for repairing DNA from formalin-fixed paraffin-embedded (FFPE) have emerged; however, a direct comparison of FFPE DNA repair methods prior to analysis on genome-wide methylation array to matched FF tissues has not been conducted.
Methods
We conducted a systematic performance comparison of two DNA repair methods (REPLI-g Ligase vs. Infinium HD Restore Kit) on FFPE-DNA compared to matched FF tissues on the Infinium 450K array. A threshold of discordant methylation between FF-FFPE pairs was set at Δβ>0.3. The correlations of β-values from FF-FFPE pairs were compared across methods and experimental conditions.
Results
The Illumina Restore kit outperformed the REPLI-g ligation method with respect to reproducibility of replicates(R2>0.970), highly correlated β-values between FF-FFPE(R2>0.888), and fewest discordant loci between FF-FFPE(≤0.61%). The performance of the Restore kit was validated in an independent set of 121 FFPE tissues.
Conclusions
The Restore kit outperformed RELPI-g ligation in restoring FFPE-derived DNA prior to analysis on the Infinium 450K methylation array. Our findings provide critical guidance that may significantly enhance the breadth of diseases that can be studied by methylomic profiling.
Impact
Epigenomic studies using FFPE tissues should now be considered among cancers that have not been fully characterized from an epigenomic standpoint. These findings promote novel epigenome-wide studies focused on cancer etiology, identification of novel biomarkers, and developing targeted therapies.
Keywords: Genome-wide methylation array, epigenomics, cancer, formalin-fixed paraffin-embedded tissue, DNA repair
INTRODUCTION
Epigenome-wide association studies (EWAS) (1) focused on disease risk (2, 3) and environmental exposures (4, 5) are emerging in the field of cancer epidemiology. DNA methylation is a stable epigenetic modification of DNA that occurs primarily at cytosine-guanine (CpG) dinucleotide pairs within CpG islands in close proximity to gene promoters (6). DNA methylation alterations are critical events in carcinogenesis that parallel genomic mutational alterations and often occur early in carcinogenesis (7–10). Genome-wide methylation profiling is technically feasible with the emergence of several large-scale methylation assays, including the Illumina Infinium 450K arrays (11, 12), methylated DNA immunoprecipitation (Me-DIP) (13), and reduced representation bisulfite sequencing (RRBS) (14). The Infinium 450K array is a high-throughput assay interrogating >450,000 CpG-sites and considered the platform best suited for large studies (11, 12). However until recently, its use had been limited to high-quality high-molecular weight DNA (11) derived from flash-frozen (FF) tissues that are uniformly fragmented for unbiased whole genome amplification (WGA) (15). Due to the formalin-induced damage (e.g. non-uniform fragmentation and cross-linking) (16), DNA from formalin-fixed paraffin-embedded (FFPE) tissues has been generally unsuitable for WGA due to potential biased amplification (15). This limitation of the methylation array technologies has limited molecular epidemiological studies focused on the epigenome of cancers for which only FFPE blocks are available.
Overcoming the technical limitations inherent to genome-wide methylation array of FFPE specimens is critical to leverage the power of epigenomic analyses for many cancers and to promote EWAS (1). To address the DNA requirements for the Infinium platform, Thirwell et al. proposed a novel method to repair random DNA fragmentation by random ligation (REPLI-g ligase protocol) prior to bisulfite modification. This method generated highly concordant methylation values (e.g. R2=0.97) from paired FF-FFPE tissues using the Illumina 27K array (17), which overall were replicated by Jasmine et al. (18). However, the performance of the Thirwell technique of FFPE-DNA (e.g. a comparison of paired FF-FFPE tissues) prior to analysis on the Infinium 450K array, which interrogates a larger portion of the genome, is unknown. In addition, Illumina developed the Infinium HD FFPE Restore Kit (19), which repairs bisulfite-modified FFPE DNA prior to WGA specifically for the Infinium 450K array using a combination of DNA polymerases and ligases to restore DNA length. The Restore protocol differs from Thirwell’s protocol with respect to (1) the order of ligation and bisulfite modification, (2) the ligase/polymerase methodology and (3) the minimum starting DNA requirements. Furthermore, the Thirwell REPLI-g ligase method is less expensive and requires less processing time; therefore, may be an optimal choice for large-scale population studies. However, the performance of the Restore kit has not been directly compared to the REPLI-g ligase for DNA interrogated by the Illumina 450K array. Herein, we report an independent and systematic comparison of the REPLI-g Ligase method and Ilumina Restore Kit for repairing FFPE-DNA prior to WGA and analysis on the Infinium 450K array. We examined several parameters that differed between the methods as well as several data normalization methods. The optimal method was successfully tested on 121 archived FFPE tissues from a completed national clinical trial, Radiation Therapy Oncology Group (RTOG) trial 98-11.
MATERIAL AND METHODS
Overview of Experimental Parameters
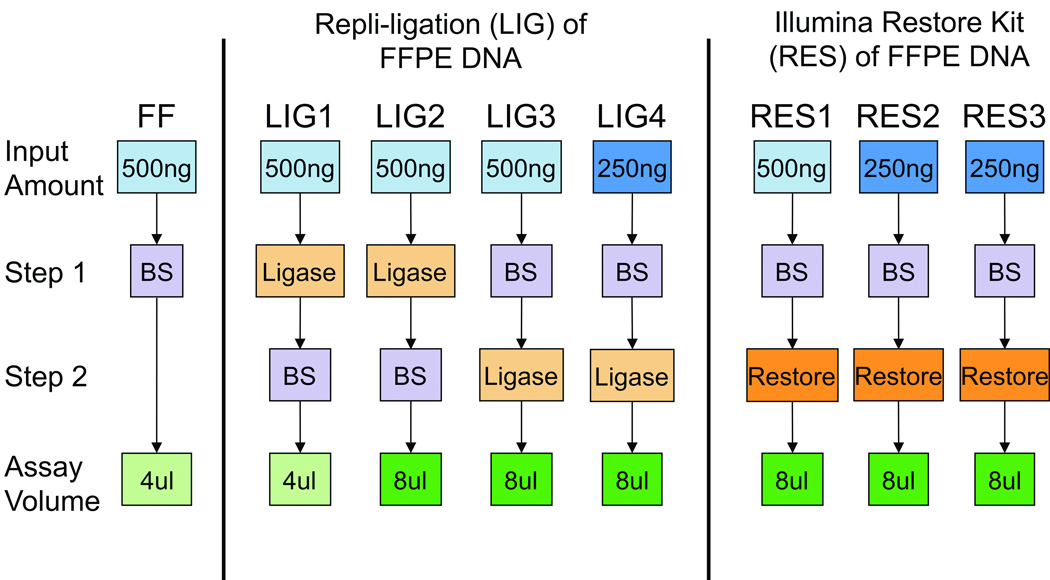
The REPLI-g ligase (LIG) and Illumina Restore (RES) methods for repairing DNA extracted from FFPE blocks were compared, as outlined in Figure 1. The original Thirwell method (17) was tested using 500ng of genomic DNA that underwent REPLI-g ligation and bisulfite modification with 4µl (LIG1) or 8µl (LIG2) of bisulfite-modified DNA used for WGA and Illumina 450K array. LIG1 and LIG2 were considered REPLI-g ligase technical replicates. Bisulfite modification degrades genomic DNA (20) and may counteract the function of the ligase when performed after ligation; therefore, bisulfite modification of 500ng (LIG3) or 250ng (LIG4) of genomic DNA was conducted prior to REPLI-g ligation. The minimum starting amount of DNA is 500ng from FF and 250ng (19) or 500ng (17) from FFPE, depending on the repair method. DNA inputs of 500ng (RES1) and 250ng (RES2/RES3) were used for the Illumina Restore kit. RES2 and RES3 were technical replicates for the Illumina Restore method. DNA from RTOG tissues was processed according to RES2 parameters (Figure 1).
Figure 1. Schematic of Experimental Parameters comparing FFPE DNA repair methods to FF DNA.
DNA (500ng) from FF tissue was processed according to Illumina Human Methylation instructions, including bisulfite modification followed by the standard Human Methylation processing protocol (Gold standard). Experimental conditions are separated for REPLI-g ligation (LIG) and Illumina Restore Kit (RES). LIG1: the original Thirwell method using 500ng of genomic DNA processed by REPLI-g ligase and bisulfite modified (BS), and 4µl of bisulfite-modified DNA used for the starting material for the Illumina Human Methylation array kit. LIG2: Thirwell method with output DNA increased to 8µl of bisulfite modified DNA, which is the same as used in the Restore Kit. LIG3 and LIG4: 500ng and 250ng of genomic DNA, respectively, were bisulfite modified, processed by REPLI-g ligase and 8µl of material used for Illumina Methylation Array kit. RES1: the Illumina Restore protocol using 500ng of genomic DNA, bisulfite modified and processed per Restore Kit protocol (including 8µl for Array steps). RES2 and RES3: Technical replicates for Restore Kit using 250ng of genomic DNA.
Tissues Specimens
FF and FFPE specimens
Matched FF colon adenocarcinoma tissue and corresponding archived FFPE blocks were obtained from three patients between 1999 and 2008 (2 males and 1 female; Supplementary Table S1). Histopathology was reviewed and confirmed by a dedicated gastrointestinal pathologist. Similar regions of invasive tumor within FF and FFPE specimens were marked and macrodissected. This study was reviewed by the University of South Florida IRB and determined to be human subject exempt research.
Archived FFPE Tissues from Radiation Therapy Oncology Trial (RTOG) 98-11
As a validation set, we utilized archived FFPE sections collected in the RTOG 98-11 trial for the treatment of squamous cell carcinoma of the anus (21). Sections from archived FFPE tissues (4×10 microns section or 8×5 micron sections) were obtained from 186 cases, pathologically reviewed and macrodissected (22).
DNA Extraction
DNA from FF tissue was isolated using the QIAamp DNA Blood Mini Kit (QIAGEN Inc., Valencia, CA) following manufacturer’s recommendations. DNA from sections of colon FFPE blocks (e.g. 10 microns × 8–10 slides) and RTOG sections was isolated using the QIAamp DNA FFPE Tissue Kit (QIAGEN Inc., Valencia, CA). DNA was evaluated using the NanoDrop1000 Spectrophotometer (Wilmington, DE) and double-stranded DNA concentration determined using the Qubit® 2.0 Fluorometer.
Quality Control
Quality of FFPE DNA was tested in triplicate by real-time PCR using the Illumina FFPE QC kit (Illumina, Inc., San Diego, CA). Amplification of the FFPE sample DNA was compared to the amplification of a Quality Control template (QCT). The real-time PCR threshold cycle (Ct), or the quantification value, was averaged across replicates and a ΔCt for each sample was calculated (CtFFPE − CtQCT). An FFPE DNA sample was deemed adequate for the Illumina 450k array if the ΔCt was <5.
Bisulfite Conversion
Genomic DNA was directly subjected to bisulfite modification for FF samples, LIG3 and LIG4, and all RES samples; whereas DNA for LIG1 and LIG2 was processed using the REPLI-g ligase prior to bisulfite modification (Figure 1). Genomic DNA (500ng or 250ng) or REPLI-g ligated DNA in 45µl volume was bisulfite modified using the EZ DNA Methylation Kit (cat#D5001, Zymo Research, Irvine, CA) following manufacturer's instructions. A volume of 9.5µl M-elution buffer was used for all conditions except LIG3 and LIG4, which required 11.5µl M-elution buffer.
REPLI-g Ligation of FFPE Samples
The REPLI-g FFPE Kit (QIAGEN Inc., Valencia, CA) was tested following Thirwell’s protocol (17) for LIG1 and LIG2. The sequence of bisulfite modification was altered for LIG3 and LIG4, with bisulfite modified DNA treated with REPLI-g ligase following published protocol followed by the Zymo Clean & Concentrator kit (Zymo Research Corporation, Irvine, CA) prior to processing for the Illumina 450K array (LIG 3 and LIG4).
Restoration of FFPE Samples
Infinium HD FFPE DNA Restore kit (Illumina, Inc., San Diego, CA) (19) protocol was carried out according to the manufacturer’s instructions on 8µl of bisulfite-treated FFPE DNA (RES1, RES2, & RES3). The DNA was eluted with DiH20 after a 5 minute incubation and stored at −20°C prior to the Infinium processing.
Genome-wide Methylation Assay
The Infinium Human Methylation 450K Beadchip® (Illumina, Inc., San Diego, CA) measures DNA methylation at 482,421 CpG loci, which covers ~99% of RefSeq genes, 96% of UCSC CpG islands and specific commonly methylated CpG sites in human cancers (www.ncbi.nlm.nih.gov/RefSeq, www.illumina.com). This protocol was carried out according to the manufacturer’s recommendations. All samples utilized 8 µl starting material except for FF and LIG1, which used 4µl of bisulfite-modified DNA as per Thirwell (17). A Tecan Liquid Handling robot with the Te-Flow apparatus was used for the single base extension and staining, and the chips were scanned on a single HiScanSQ System (Illumina Inc.). Paired FF and FFPE DNA were processed on the same chip to reduce batch effects. RTOG DNA was run in 3 batches of 24 and included two interbatch technical replicates (Case ID 37 and 370).
Data Analysis
Three data normalization methods [minfi package-implemented Illumina (23), SWAN (24), DASEN (25)] and raw β-values were examined. The β-value is calculated for the 485,512 CpG-loci from unmethylated (U) and methylated (M) signal [M / (U+M+100)] and assigned a range between 0–1 (unmethylated to 100% methylated) within the Illumina Infinium software and provided within the raw idat file. The raw idat files were processed for all normalization packages in R/Bioconductor (26). The minfi Bioconductor package (23) (version 1.8.7) generated three distinct MethylSet objects: 1) the preprocess-Raw method, 2) preprocess-Illumina method with background correction and normalization via controls, and 3) preprocess-SWAN method (24). β-values for each MethylSet were extracted using minfi’s getBeta method with the parameter type = “Illumina” to ensure that the β-values were calculated with an offset of 100. The DASEN normalization used the DASEN method in the WateRmelon package (27) (version 1.2.2) (25). All normalizations ranked the quality of experimental conditions identically; primary results are reported using the DASEN normalization method.
Normalized β-values for CpG loci with a detection p-value>0.05 were removed. β-values were analyzed as continuous variables. To quantitate the discordant β-values between FF DNA and FFPE repaired DNA, we calculated a Δβ = β-valueFF − β-valueFFPE. The number of CpG loci with Δβ either above or below (e.g. absolute value of Δβ noted as |Δβ|) 0.3 was calculated. Correlation between FF and FFPE was determined using Pearson correlation coefficient. Significant differences in mean R2 or Δβ between REPLI-g Ligase and Restore methods were determined using a paired t-test (two-sided). All calculations and correlations were done in MATLAB. Principal component analysis (PCA) was performed using Evince v2.5.5 software (UmBio AB, Umeå, Sweden, www.umbio.com).
RESULTS
We examined two methods [REPLI-g ligase (LIG) and Illumina Restore (RES)] for repairing DNA extracted from FFPE tissues and analyzed on the Infinium 450K array. Three paired sets of FF-FFPE colon tumors that varied by time since collection (4–13 years) and gender were used (Supplementary Table S1). The experimental design and standard nomenclature are presented in Figure 1 and outlined in Materials and Methods.
Patient variability drives the largest differences in β-values
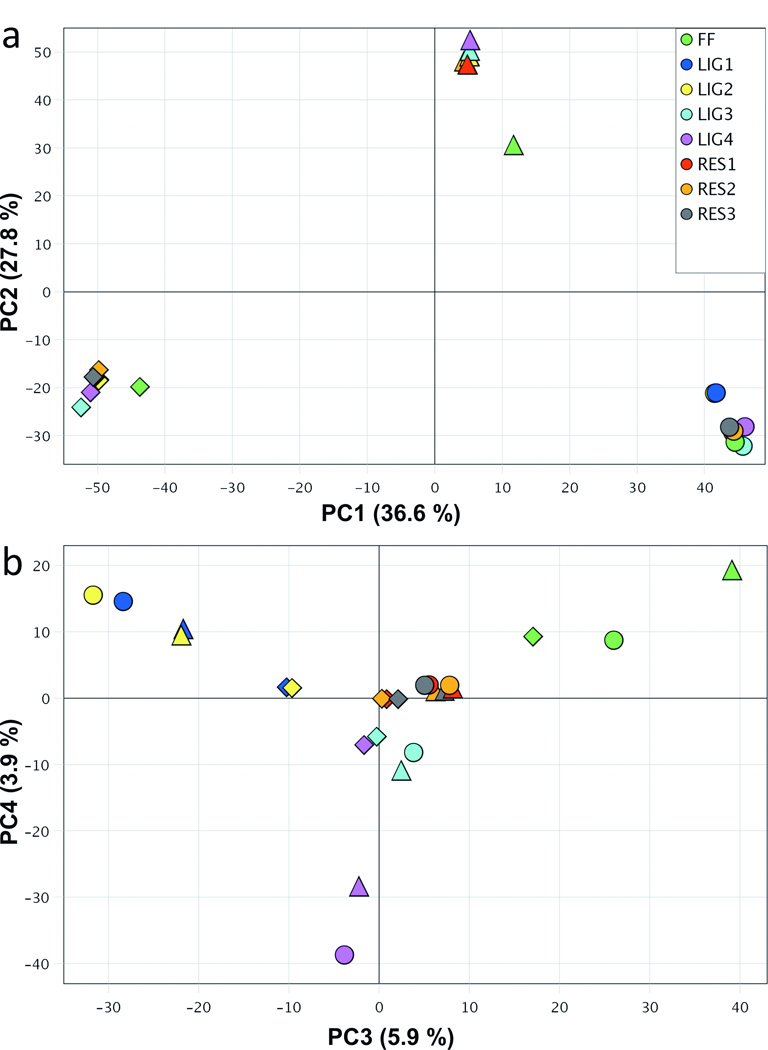
The sources of variation in methylation β-values across the patient samples and all conditions were examined using principal components analysis (PCA). As shown in Figure 2a, the samples clustered primarily by patient (triangles, circles and diamonds) in the first and second principal components (PC1 and PC2), which accounted for 36.6% and 27.8% of the variation in β-values, respectively. This indicates that patient-related differences in β-values are larger than differences by tissue storage type (FF vs. FFPE) or ligation method (LIG vs. RES). PC3 and PC4 (Fig. 2b) explain an additional 5.9% and 3.9% of the variation in β-values, respectively. There was a clear similarity among all Restore samples across patients (e.g. clustering of red, orange and gray points) in PC3 and PC4 with little variability compared to the separation of the REPLI-g ligase-processed samples (yellow, dark blue and purple points; Fig. 2b). The LIG3 samples (light blue points) were most similar to the Restore samples and clustered together across patients. In summary, inter-patient variation in β-values was high; all Restore samples and LIG3 displayed the least variation in β-values between patients and were most similar to FF samples.
Figure 2. Principal component analysis (PCA) to identify the sources of variation in methylation β-values across the three patient samples and all conditions.
Patient samples are denoted by shape (A – circle, B – triangle and C – diamond). (a) Scatter plot of samples in the first and second principal components (PC1 and PC2), which account for 36.6% and 27.8% of the variation. (b) Scatter plot of PC3 and PC4, which explained 5.9% and 3.9% of the variation with separation by tissue storage type (FF vs. FFPE) as well as the ligation method (LIG vs. RES).
Reproducibility of FFPE-based genome-wide methylation is higher using Restore method
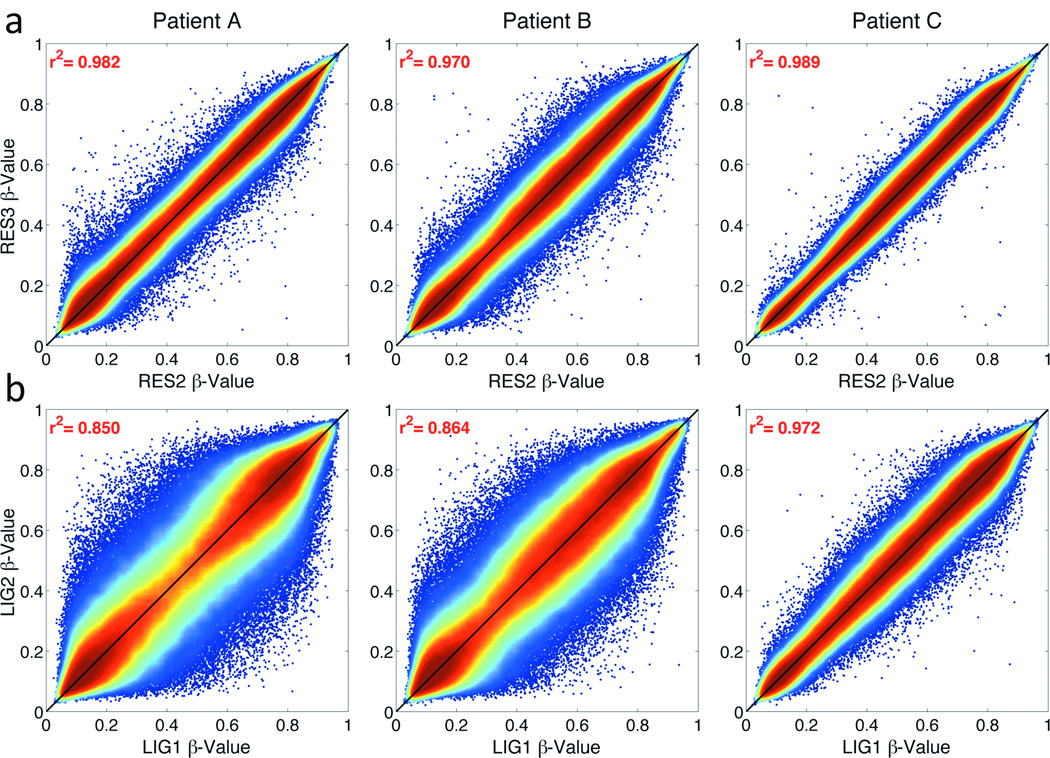
The reliability of each DNA-repair method was assessed using β-values from the Infinium 450K array of technical replicates (RES2/RES3 and LIG1/LIG2). The correlation between Restore internal replicates (RES2 vs. RES3) ranged from 0.970–0.989 (Fig. 3a). The correlations between REPLI-g ligase replicates (LIG1 vs. LIG2) were lower (0.850–0.972, Fig. 3b). REPLI-g ligation resulted in more β-values off the diagonal (Fig. 3b) compared to the Restore replicates (Fig. 3a). There were 346 total CpG loci across patients with discordant β-values (|Δβ|) of >0.3 between RES2 and RES3 (Supplementary Table S2). By comparison, REPLI-g ligase replicates had a total of 12,718 loci across patients with |Δβ|>0.3.
Figure 3. Density correlation plot between internal replicate samples.
Correlations are shown using (a) the Restore kit (RES2 vs. RES3) and (b) REPLI-g Ligase (LIG1 vs. LIG2) for each tumor. Colors range from blue (low point density) to red (high point density), with the highest density along the diagonal.
Highest correlations between FF-FFPE β-values are achieved using Restore
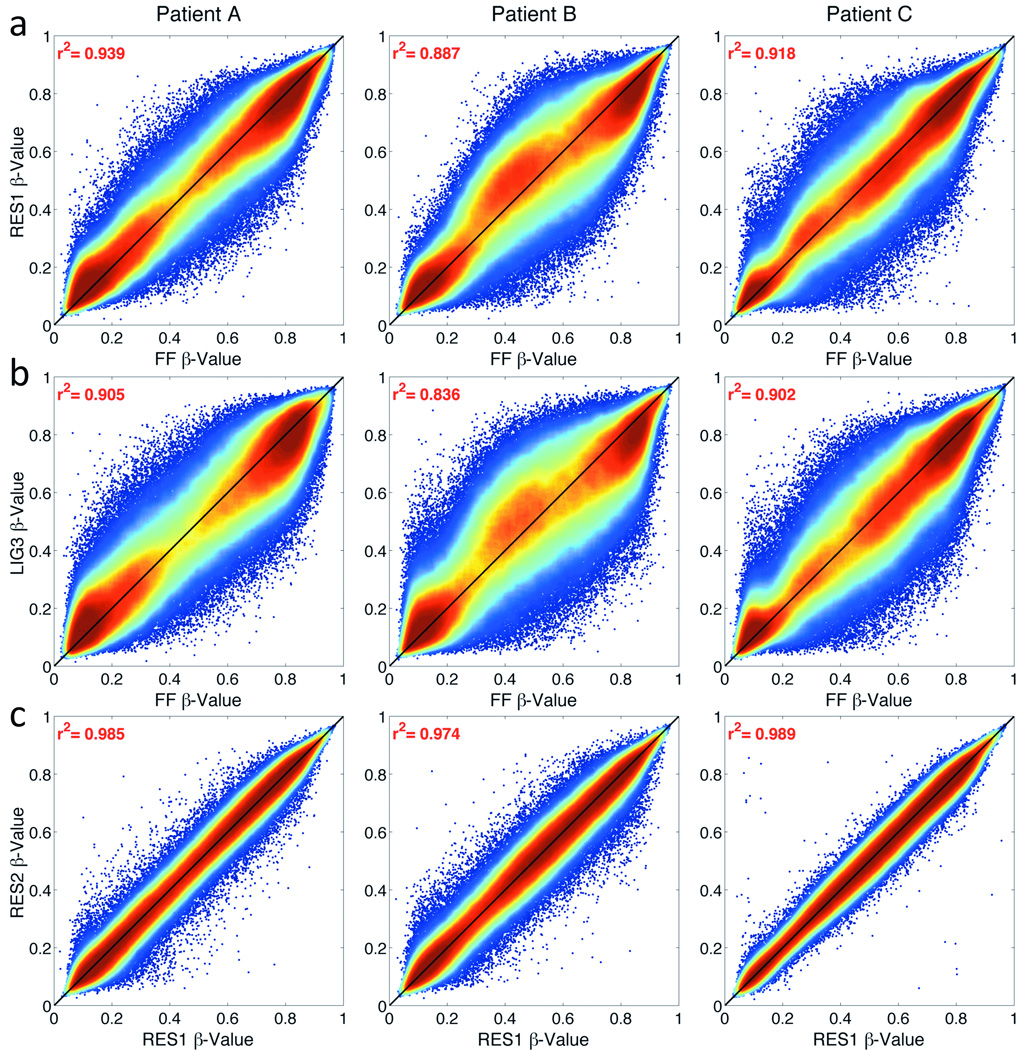
The goal of both methods is to repair DNA such that amplification bias during WGA due to potentially non-uniformly fragmented DNA from FFPE preserved tissues is reduced. To quantify if the DNA repair methods reduce this bias and methylation values from FFPE-derived DNA are representative of FF-derived DNA, the β-values from FFPE-derived DNA were compared to β-values from matched FF-derived DNA. Overall, the average correlation between FF-FFPE pairs ranged from 0.804 to 0.938 (Table 1). For all samples, the correlations between FF and the Restore-treated FFPE DNA were significantly higher than the REPLI-g ligase-treated FFPE DNA (Mean R2: 0.91 vs. 0.86, respectively, p<0.003). Correlations between FF-FFPE pairs using the Restore method were highest for RES1 (e.g. RES1 vs. FF, R2=0.888–0.938; Fig. 4a) but did not differ by DNA input amount (Table 1). Correlations using the REPLI-g ligase method were all lower than Restore. LIG3 had the highest correlations among the LIG samples (e.g. LIG3 vs. FF, R2=0.836–0.905; Fig. 4b). For Restore, β-values from 250ng and 500ng of FFPE DNA (Thirwell recommended starting amount) were highly correlated (RES1 vs. RES2, R2=0.974–0.989; Fig. 4c) and at a similar magnitude as seen between technical replicates (RES3 vs. RES2, R2=0.970–0.989; Fig. 3a).
Table 1.
Summary of β–value correlation (R2) and discordance (|Δβ|) between FF-FFPE pairs for REPLI-g Ligation and the Restore methods.
| R2 of β-values FF-FFPE pairsa | Discordant loci between FF-FFPE pairsb | Overlap of discordant locic |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Patient | Overall | Patient | Overall | % CpG- Loci |
Pairwise by Patient |
Over all |
|||||||
| A | B | C | Mean ± Std | A | B | C | Mean ± Std | AB | AC | BC | ABC | ||
| LIG1 | 0.857 | 0.812 | 0.900 | 0.856 ± 0.044 | 8,254 | 10,338 | 3,772 | 7,455 ± 3,355 | 1.54% | 638 | 120 | 137 | 23 |
| LIG2 | 0.849 | 0.810 | 0.901 | 0.853 ± 0.046 | 9,415 | 10,638 | 3,834 | 7,962 ± 3,627 | 1.64% | 728 | 139 | 142 | 27 |
| LIG3 | 0.905 | 0.836 | 0.902 | 0.881 ± 0.039 | 3,212 | 9,892 | 4,323 | 5,809 ± 3,579 | 1.20% | 190 | 65 | 156 | 7 |
| LIG4 | 0.860 | 0.804 | 0.898 | 0.854 ± 0.047 | 10,250 | 15,168 | 4,390 | 9,936 ± 5,396 | 2.05% | 774 | 163 | 244 | 19 |
| RES1 | 0.939 | 0.888 | 0.918 | 0.915 ± 0.026 | 1,124 | 3,402 | 3,206 | 2,577 ± 1,262 | 0.53% | 111 | 51 | 65 | 19 |
| RES2 | 0.941 | 0.875 | 0.919 | 0.912 ± 0.034 | 1,149 | 4,662 | 3,053 | 2,955 ± 1,759 | 0.61% | 146 | 34 | 72 | 14 |
| RES3 | 0.938 | 0.881 | 0.920 | 0.913 ± 0.029 | 1,214 | 3,969 | 3,263 | 2,815 ± 1,431 | 0.58% | 136 | 35 | 65 | 15 |
Pearson Correlation Coefficient comparing FF vs. FFPE β-values for each patient sample, by experimental condition. Mean R2 for LIG and RES samples differed significantly by Student t-test (p=0.0028)
The number of discordant CpG-loci defined as |Δβ|>0.3 between FF and FFPE. Mean number of discordant CpG-loci differed significantly between LIG and RES samples (Students t-test, p=0.0012). Percent of CpC-loci with |Δβ|>0.3 out of total CpG-loci evaluated (N=485,512) for each condition
Overlap of individual discordant CpG-loci (|Δβ|>0.3) across patient samples (A, B and C), pairwise and overall.
Figure 4. Representative density correlation plot between FF-FFPE pairs.
Correlation of DASEN normalized β-values for (a) Restore (RES1 vs. FF) and (b) REPLI-g Ligase (LIG3 vs. FF) for each patient sample. (c) Correlation between FFPE Restore-processed samples by input DNA amount (500ng RES1 vs. 250ng RES2).
Restore yields fewer CpG loci with discordant FF-FFPE β-values
Discordant β-values (|Δβ|) between FF-FFPE tissues were examined for the different techniques by determining the average number of CpG loci above a |Δβ| threshold of 0.3. The mean number of loci with |Δβ|>0.3 for RES samples ranged from 2,577 to 2,955 (0.53%–0.61% of all loci; Table 1). The mean number of loci with |Δβ|>0.3 was 2 to 3 fold higher in the LIG samples than RES (5,809–9,936 loci, 1.2%–2.05%). Restore processed DNA had significantly fewer CpG loci with |Δβ|>0.3 between FF-FFPE compared to REPLI-g ligase-processed DNA (p<0.002).
FF-FFPE β-value discordance occurs at random CpG loci
Next, it was determined whether discordant β-values occurred in the same CpG loci (differential bias) across samples or whether they occurred at random (non-differential bias) by examining the overlap in loci with |Δβ|>0.3 across patient samples and various conditions. The number of loci that were consistently discordant between FF-FFPE across all three patient samples (A&B&C) ranged from 7 to 27 (Table 1). Of note, no CpG loci had a |Δβ|>0.3 consistent across all seven experiments. These observations were consistent for Restore replicates (Supplementary Table S2), with no overlapping loci with |Δβ|>0.3 across all three patients. These results suggest that loci with discordant β-values between matched FF-FFPE DNA samples occurred at random and were not due to specific poorly performing loci in FFPE samples.
Impact of Modulating Experimental Parameters
Experimental conditions that differed across the two methods were evaluated (Fig. 1). There were no differences for Restore-processed DNA by starting DNA (500ng vs. 250ng) for either the correlations or number of loci with |Δβ|>0.3 (Fig. 4c and Table 1). For REPLI-g ligase, 500ng starting DNA (LIG3) performed better than 250ng (LIG4) for all metrics examined. LIG3 had the fewest number of loci with |Δβ|>0.3 (mean loci=5,809, 1.20% of all loci) among REPLI-g ligase experiments (Table 1) and clustered with RES samples by PCA (Fig. 2b). Bisulfite modification prior to REPLI-g ligation (LIG3/4) outperformed the reverse sequence (LIG1/2) with higher R2 (mean R2=0.867 and 0.855, respectively; Table 1). Finally, there was minimal impact of changing the volume of material taken forward for the Infinium assay (Mean |Δβ|>0.3: 7,455 and 7,962 for LIG1 and LIG2, respectively; Table 1).
Impact of data normalization
There is consensus that Infinium 450K β-values should be normalized (28, 29) and several methods have been proposed. We considered three normalization methods (minfi-Illumina, SWAN and DASEN) in addition to using raw (un-normalized) β-values. Overall, all normalization techniques ranked the different experimental conditions identically on all performance metrics. Notably, there were differences observed in the magnitude and direction of the Δβ between FF-FFPE by normalization method. Compared to DASEN (Fig. 4), there was a tendency for minfi-Illumina to overestimate the methylation status of FFPE samples (vs. FF) with substantially more loci above the diagonal line (Supplementary Figure S1). The Δβ-values between FF-FFPE samples were normally distributed with a mean Δβ of 0.002 using the DASEN method. There was a significant shift toward higher β-values within the FFPE samples (negative Δβ) when using minfi-Illumina (mean Δβ= −0.028, p=6.03×10−8) (Supplementary Figure S2). Among all methods, DASEN also yielded significantly fewer loci with |Δβ|>0.3 (mean |Δβ|>0.3=7,831 across LIG samples and 2,794 for RES) while minfi-Illumina resulted in the highest number (mean |Δβ|>0.3 = 18,082 across LIG samples and 5,528 for RES, p<0.0001). The mean Δβ and number of discrepant loci (|Δβ|>0.3) between FF-FFPE for all normalization methods are presented in Supplementary Tables S3 and S4, respectively.
Testing of Restore methodology in archived FFPE-tissues
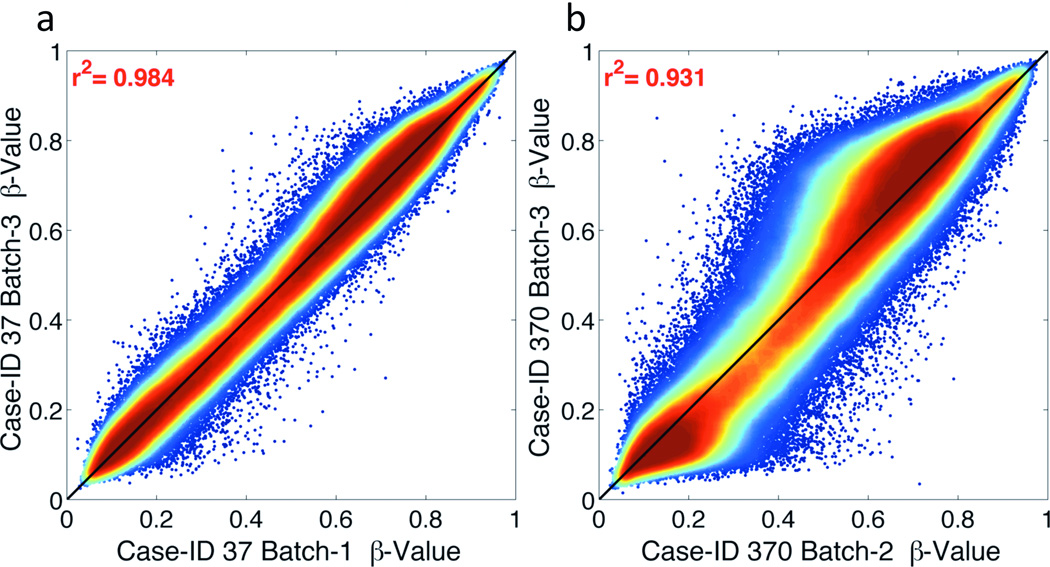
The optimized conditions of 250ng FFPE-derived DNA using the Illumina Restore kit (RES2) were applied to a set of archived FFPE tissues collected between 1998 and 2005 as part of a cooperative group anal cancer clinical trial. Of the 186 cases, 121 (65%) had ≥250ng DNA. In QC testing, the ΔCt of DNA from RTOG FFPE tissues ranged from −1.03 to 4.73; all 121 samples passed QC testing. Percent missing CpG loci were <3% for all but 1 sample (>5%), which was excluded. β-values from two replicate samples repeated across batches (Case-ID 37 and 370) were highly correlated (R2=0.984 and 0.931, Fig. 5) and had few CpG loci with |Δβ|>0.3 (29 and 1,483, respectively). The distribution of β-values for the 121 FFPE samples from RTOG exhibited a consistent pattern (Supplemental Fig. S3b) that was comparable to FF-FFPE pairs (Patient A, B and C, Supplemental Fig. S3a). All samples, regardless of storage type, had β-value peaks at 0.2 and 0.8 and variability between β-values of 0.3 and 0.6. PCA for all samples showed no outliers or batch-effects (data not shown). Taken together, these data demonstrate that using the Illumina Restore Kit with 250ng of genomic DNA from archived pre-treatment biopsy specimens >10 years-old can result in high quality epigenomic data.
Figure 5. Density correlation plots for RTOG FFPE tissue internal replicates using Restore method.
Correlations are shown using the Restore kit (RES2 condition) for archived FFPE tissues collected from Case-ID 37 (a) and 370 (b) within the RTOG 98-11 clinical trial. Colors range from blue (low point density) to red (high point density), with the highest density along the diagonal as expected.
DISCUSSION
The Illumina Restore kit outperformed the RELPI-g ligation method for restoring FFPE-derived DNA prior to use on the Infinium 450K methylation array. The Restore method had the best overall performance regardless of starting amount of DNA tested and was consistent across several data normalization methods, although DASEN normalization performed best. The drawbacks of the Restore kit include added costs per sample ($80 vs. $24) and additional processing time (~4 hours per-batch). The Thirwell method with modification may also be an acceptable option when sufficiently large effect sizes are expected (e.g. |Δβ|>0.5). Our findings provide valuable guidance for selecting a DNA repair method for FFPE samples prior to analysis on the Infinium 450K array and highlights several important factors for consideration when designing epigenome-wide association studies, including cost, DNA requirements, and processing of resultant data.
Many cancers, such as anal, rectal, and esophageal, are treated with chemoradiation prior to surgery and only small pre-treatment diagnostic FFPE biopsies are available for molecular analysis. The best method for repairing DNA from these tissue types for genome-wide methylation analysis (e.g. most representative of matched FF tissue) had not been determined. Our study provided evidence that “restored” FFPE-derived DNA generated β-values from the Illumina 450K array that were representative of FF tissues. We then validated these finding using DNA from pre-treatment FFPE biopsies archived as part of the RTOG 98-11 anal cancer trial (21). DNA extracted from pre-treatment FFPE biopsies met quality standards and yielded high-quality methylation data, as determined by a low percentage of undetectable probes, a consistent distribution of β-values across the array, and highly correlated β-values from sample replicates. Notably, the use of 250ng starting DNA, instead of 500ng as recommended by Thirwell, increased our sample size by 43 cases (36% increase) with DNA yields between 250ng and 500ng. The delineation of the optimal experimental conditions and restore methods should open the door for the characterization of genome-wide methylation in a number of cancers that remain heretofore inadequately studied from an epigenomic standpoint.
Using a comprehensive set of quality measures, we systematically compared two methods and three experimental parameters for repairing non-uniformly fragmented FFPE-derived DNA prior to WGA and evaluation on the Infinium 450K methylation array. To our knowledge, this is the first study to directly compare the Illumina Restore kit to the methods by Thirwell et al. (17) prior to analysis on the Infinium 450K array. The overall correlation of β-values between FF and FFPE tissues observed in this study was similar, if not greater, than those previously reported for the Infinium 27K array (17, 18). Lechner et al. demonstrated that FFPE DNA repaired using the REPLI-g ligase was sufficient for use on the Illumina 450k array when stringent QC criteria were applied prior to data analysis and large effect sizes were observed (e.g. differential methylation between human papillomavirus (HPV) positive vs. HPV negative head and neck cancers) (30). In contrast to Jasmine et al. (18), our data suggest that there is minimal misclassification of β-values generated from FFPE DNA for both the REPLI-g ligase and the Restore; however, the Restore method outperformed the REPLI-g ligase method.
Overall, our findings that the Restore processed samples provided β-values representative of FF tissues are in agreement with those reported by Dumenil et at (31), who analyzed 21 FF-FFPE colorectal cancer tissue pairs using the Illumina Restore and Illumina 450K array. They reported highly consistent β-values across FF-FFPE pairs, very low percentage of undetectable probes (<1%), and overlap in differentially methylated loci between FFPE and FF tissues (31). However, Dumenil et al. did not compare the Restore method to the less-expensive REPLI-g ligation method nor did it consider more than one data normalization method. As both our study and Dumenil observed some differences in β-values between FF and restored FFPE-derived DNA (as expected), we strongly advocate that a minimum |Δβ|-threshold (e.g. |Δβ|>0.3) be utilized, in addition to statistical significance, when identifying differentially methylated loci from Illumina 450K data.
The array-based DNA methylation data normalization field is still nascent with new normalization techniques being proposed and methods continuing to be debated (see Wilhelm-Benartzi et al. (28) for review). This is in contrast to more mature array-based methods such as gene expression. An important observation of this study is that the selection of the best experimental protocol was independent of the four different normalization methods investigated. Nonetheless, the DASEN (25) technique performed best among the methods tested. This could be explained by the fact that DASEN is a global sample-to-sample normalization technique while minfi-Illumina and SWAN normalize each sample independently.
The validity of our findings is strengthened by our experimental rigor including the inclusion of all paired samples on the same chip to reduce chip-to-chip variation. Although our experimental study consisted of only three paired cases, these specimens are representative of typical FFPE tissues that would be included in larger studies with variation in specimen age and patient gender. This was also evident by the variability observed in the methylation data. The experimental findings were successfully tested on 121 anal cancer FFPE tissues collected within a clinical trial; thus demonstrating the applicability of this method to clinical specimens from several pathology laboratories. We included two types of cancer in this study, colorectal and anal to represent (1) tumors with sufficient material to obtain FF and FFPE matched samples that provided >2ug of DNA and (2) small pre-treatment biopsies with minimal amounts of DNA, respectively. This study did not include normal tissues, as examined by Jasmine et al. (18) However, as the goal was to identify a method that generated methylation results within FFPE tissues as similar as possible to matched FF tissue, the lack of normal tissues or use of different tumor types did not impact our conclusions.
This study demonstrates that FFPE derived DNA processed using the Illumina Restore provides robust genome-wide methylation results that are similar to those from optimally stored matched FF tissues. This DNA repair method is recommended above the REPLI-g ligation method for any future epigenomic studies. Aberrant methylation occurs during critical processes of aging, development and carcinogenesis (2, 3); as such, these recommendations will have widespread implications and should greatly increase the breadth of diseases that can undergo methylomic profiling. This may, in turn, lead to further elucidation of disease pathogenesis, identification of novel biomarkers, and the development of targeted therapies.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Thomas Sellers and Dr. Alvaro Monteiro for their critical review of this paper and Dr Bill Grady and Dr. Andrew Kaz for critical review of our study design. We are grateful for our ongoing collaboration with the Radiation Therapy Oncology Group 98-11 Investigators (Drs. Jaffer Ajani and Chandan Guha) and Statisticians (Ms. Kathryn Winters and Jen Moughan) in the use of RTOG 98-11 tissue and data.
FUNDING: The research was supported in part by the American Society of Colon and Rectal Surgeons Foundation, under grant number LPG-097 (EM Siegel, D Shibata). This work has been supported in part by the Cancer Informatics and Molecular Genomics Core Facility at the H. Lee Moffitt Cancer Center & Research Institute, an NCI designated Comprehensive Cancer Center, under grant number P30-CA76292. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or the H. Lee Moffitt Cancer Center & Research Institute.
Footnotes
CONFLICT OF INTEREST: The authors have no conflicts of interest to disclose.
REFERENCES
- 1.Michels KB, Binder AM, Dedeurwaerder S, Epstein CB, Greally JM, Gut I, et al. Recommendations for the design and analysis of epigenome-wide association studies. Nature methods. 2013;10:949–955. doi: 10.1038/nmeth.2632. [DOI] [PubMed] [Google Scholar]
- 2.Rakyan VK, Down TA, Balding DJ, Beck S. Epigenome-wide association studies for common human diseases. Nat Rev Genet. 2011;12:529–541. doi: 10.1038/nrg3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Feinberg AP. Genome-scale approaches to the epigenetics of common human disease. Virchows Arch. 2010;456:13–21. doi: 10.1007/s00428-009-0847-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Feinberg AP. Phenotypic plasticity and the epigenetics of human disease. Nature. 2007;447:433–440. doi: 10.1038/nature05919. [DOI] [PubMed] [Google Scholar]
- 5.Besingi W, Johansson A. Smoke-related DNA methylation changes in the etiology of human disease. Hum Mol Genet. 2014;23:2290–2297. doi: 10.1093/hmg/ddt621. [DOI] [PubMed] [Google Scholar]
- 6.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 7.Timp W, Feinberg AP. Cancer as a dysregulated epigenome allowing cellular growth advantage at the expense of the host. Nature reviews Cancer. 2013;13:497–510. doi: 10.1038/nrc3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brait M, Sidransky D. Cancer epigenetics: above and beyond. Toxicol Mech Methods. 2011;21:275–288. doi: 10.3109/15376516.2011.562671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Taby R, Issa JP. Cancer epigenetics. CA Cancer J Clin. 2010;60:376–392. doi: 10.3322/caac.20085. [DOI] [PubMed] [Google Scholar]
- 10.Sjoblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 11.Bibikova M, Le J, Barnes B, Saedinia-Melnyk S, Zhou L, Shen R, et al. Genome-wide DNA methylation profiling using Infinium(R) assay. Epigenomics. 2009;1:177–200. doi: 10.2217/epi.09.14. [DOI] [PubMed] [Google Scholar]
- 12.Dedeurwaerder S, Defrance M, Calonne E, Denis H, Sotiriou C, Fuks F. Evaluation of the Infinium Methylation 450K technology. Epigenomics. 2011;3:771–784. doi: 10.2217/epi.11.105. [DOI] [PubMed] [Google Scholar]
- 13.Mohn F, Weber M, Schubeler D, Roloff TC. Methylated DNA immunoprecipitation (MeDIP) Methods Mol Biol. 2009;507:55–64. doi: 10.1007/978-1-59745-522-0_5. [DOI] [PubMed] [Google Scholar]
- 14.Gu H, Smith ZD, Bock C, Boyle P, Gnirke A, Meissner A. Preparation of reduced representation bisulfite sequencing libraries for genome-scale DNA methylation profiling. Nat Protoc. 2011;6:468–481. doi: 10.1038/nprot.2010.190. [DOI] [PubMed] [Google Scholar]
- 15.Bosso M, Al-Mulla F. Whole genome amplification of DNA extracted from FFPE tissues. Methods Mol Biol. 2011;724:161–180. doi: 10.1007/978-1-61779-055-3_11. [DOI] [PubMed] [Google Scholar]
- 16.Gilbert MT, Haselkorn T, Bunce M, Sanchez JJ, Lucas SB, Jewell LD, et al. The isolation of nucleic acids from fixed, paraffin-embedded tissues-which methods are useful when? PloS one. 2007;2:e537. doi: 10.1371/journal.pone.0000537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thirlwell C, Eymard M, Feber A, Teschendorff A, Pearce K, Lechner M, et al. Genome-wide DNA methylation analysis of archival formalin-fixed paraffin-embedded tissue using the Illumina Infinium HumanMethylation27 BeadChip. Methods. 2010;52:248–254. doi: 10.1016/j.ymeth.2010.04.012. [DOI] [PubMed] [Google Scholar]
- 18.Jasmine F, Rahaman R, Roy S, Raza M, Paul R, Rakibuz-Zaman M, et al. Interpretation of genome-wide infinium methylation data from ligated DNA in formalin-fixed, paraffin-embedded paired tumor and normal tissue. BMC research notes. 2012;5:117. doi: 10.1186/1756-0500-5-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Illumina. Infinium HD FFPE Restore Protocol, Catalog # WG-901-2004. [cited 2012 06/20/2012];2011 Available from: http://supportres.illumina.com/documents/myillumina/5c3d90a3-793c-4a8b-932b-0434590f98ef/infinium_ffpe_sample_restore_booklet_15014614_c.pdf. [Google Scholar]
- 20.Grunau C, Clark SJ, Rosenthal A. Bisulfite genomic sequencing: systematic investigation of critical experimental parameters. Nucleic Acids Res. 2001;29:E65–E65. doi: 10.1093/nar/29.13.e65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ajani JA, Winter KA, Gunderson LL, Pedersen J, Benson AB, 3rd, Thomas CR, Jr, et al. Fluorouracil, mitomycin, and radiotherapy vs fluorouracil, cisplatin, and radiotherapy for carcinoma of the anal canal: a randomized controlled trial. JAMA. 2008;299:1914–1921. doi: 10.1001/jama.299.16.1914. [DOI] [PubMed] [Google Scholar]
- 22.Siegel EM, Eschrich SA, Winter KA, Riggs B, Berglund A, Ajidahun A, et al. Epigenomic Characterization of Locally Advanced Anal Cancer: An RTOG 98-11 Specimen Study. Dis Colon Rectum. 2014 doi: 10.1097/DCR.0000000000000160. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hansen K, Aryee M. Minfi: Analyze Illumina, 450k Methylation Arrays. R package version 12. 2012 [Google Scholar]
- 24.Maksimovic J, Gordon L, Oshlack A. SWAN: Subset-quantile within array normalization for illumina infinium HumanMethylation450 BeadChips. Genome Biol. 2012;13:R44. doi: 10.1186/gb-2012-13-6-r44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pidsley R, Wong CC, Volta M, Lunnon K, Mill J, Schalkwyk LC. A data-driven approach to preprocessing Illumina 450K methylation array data. BMC genomics. 2013;14:293. doi: 10.1186/1471-2164-14-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schalkwyk LC, Pidsley R, Wong CC, wfcbN T, Defrance M, Teschendorff AE, et al. wateRmelon: Illumina 450 methylation array normalization and metrics. R package. 1.4.0. ed. 2013 [Google Scholar]
- 28.Wilhelm-Benartzi CS, Koestler DC, Karagas MR, Flanagan JM, Christensen BC, Kelsey KT, et al. Review of processing and analysis methods for DNA methylation array data. Br J Cancer. 2013;109:1394–1402. doi: 10.1038/bjc.2013.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu MC, Joubert BR, Kuan PF, Haberg SE, Nystad W, Peddada SD, et al. A systematic assessment of normalization approaches for the Infinium 450K methylation platform. Epigenetics : official journal of the DNA Methylation Society. 2014;9:318–329. doi: 10.4161/epi.27119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lechner M, Fenton T, West J, Wilson G, Feber A, Henderson S, et al. Identification and functional validation of HPV-mediated hypermethylation in head and neck squamous cell carcinoma. Genome medicine. 2013;5:15. doi: 10.1186/gm419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dumenil TD, Wockner LF, Bettington M, McKeone DM, Klein K, Bowdler LM, et al. Genome-wide DNA methylation analysis of formalin-fixed paraffin embedded colorectal cancer tissue. Genes Chromosomes Cancer. 2014;53:537–548. doi: 10.1002/gcc.22164. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.