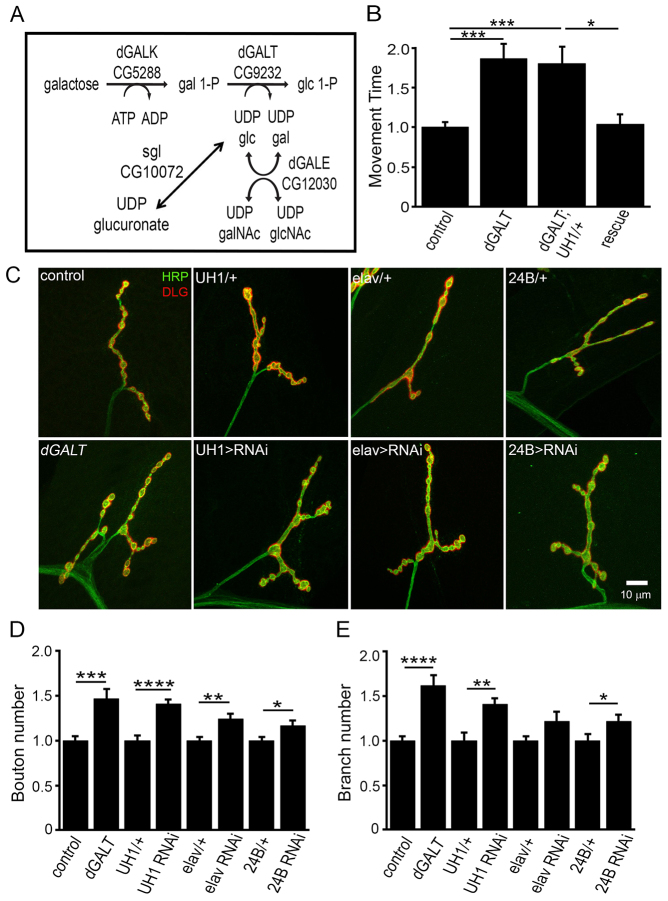
Fig. 1.
Loss of dGALT impairs coordinated movement and disrupts NMJ architecture. (A) Schematic diagram of the Leloir pathway. Galactose is phosphorylated by Drosophila galactokinase (dGALK) and then Drosophila galactose-1-P uridylyltransferase (dGALT) catalyzes the synthesis of glucose-1-P (glc-1-P) and UDP-galactose (UDP-gal) from UDP-glucose (UDP-glc) and galactose-1P (gal-1-P). UDP-glucose is a substrate for UDP-glucose dehydrogenase [encoded by sugarless (sgl)], catalyzing conversion to UDP-glucuronate, essential for proteoglycan biosynthesis. dGALE, uridine diphosphate galactose-4-epimerase. (B) Fold differences in the time required (controls set at 1) for wandering L3 to rollover from inverted to upright position for control (dGALTC2), dGALT null (dGALTΔAP2), transgenic rescue (dGALTΔAP2; UH1-Gal4/UAS-hGALT) and rescue control (dGALTΔAP2; UH1-Gal4/+). Data are normalized to respective control. (C) Representative NMJs imaged with anti-horseradish-peroxidase (HRP; green) and anti-Discs-large (DLG; red) in wandering L3 in dGALT mutants (bottom row) with complete loss (dGALTΔAP2), ubiquitous (UH1-Gal4 driven) and tissue-specific (i.e. elav- or 24-Gal4-driven) knockdown. Respective controls for each condition are shown in the top row. (D,E) Quantification of differences in NMJ bouton (D) and branch (E) number for all genotypes, normalized to appropriate controls. Sample size: ≥ten animals per genotype. Error bars show s.e.m. with significance indicated: *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001.