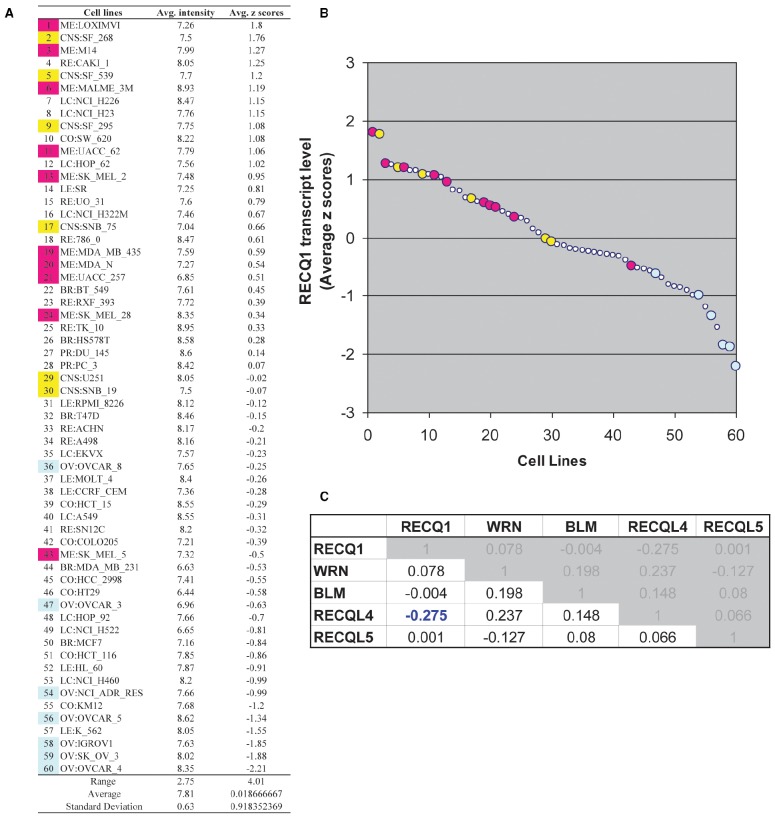
FIGURE 1.
RECQ1 transcript levels in the NCI-60 human tumor cell lines. (A) Table of average intensity (determined from log 2 intensity values from Affymetrix microarrays) and their combined z-score means presented in descending order. (B) Scatter plot depicting the distribution of RECQ1 transcript levels across the NCI-60. The numerical series (1-to-60) for the cell lines from panel (A) correspond to the x-axis. Cell lines from the central nervous system (shown in yellow) and melanoma (shown in pink) displayed higher transcript levels of RECQ1, while ovarian cancer cell lines (shown in blue) displayed the least. (C) Cross-correlation among RecQ family. Pearson’s correlation coefficients between transcripts of the known five RecQ homologs in the NCI-60. Only significant correlation of RECQ1 in the NCI-60 is with the RECQL4 transcripts. Processing and normalization of transcript expression data from the NCI-60 has been described previously (Reinhold et al., 2012). Normalization of Affymetrix HG-U95, HG-U133, HG-U133 plus 2.0 and Affy HuEx 1.0 is done by GC robust multi-array average (GCRMA). Data is accessible at http://discover.nci.nih.gov by using RECQL (RECQ1) as input in Cellminer.