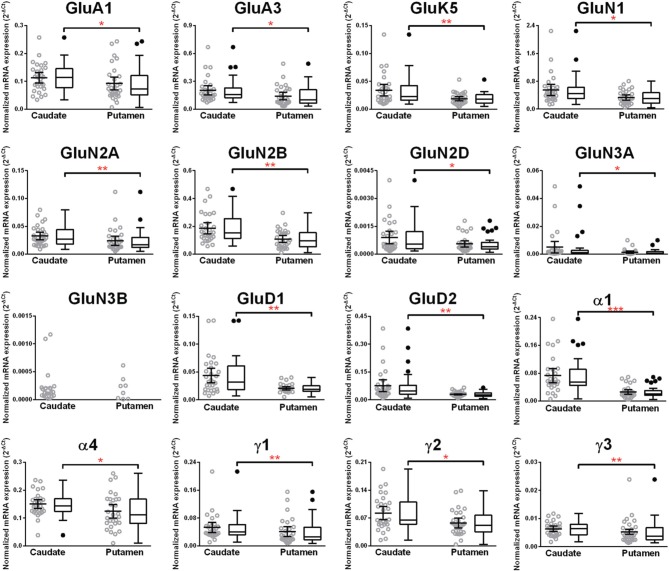
Figure 9.
Comparison of GABA-A and ionotropic glutamate receptor subunit mRNAs in the caudate and putamen of controls (n = 29). Data from each group is presented as scatter dot plot (°) with mean and 95% confidence interval and box and whiskers plot with median and whiskers plotted by Tukey method to determine outliers (•, above or below the whiskers). Statistical analysis was performed by excluding outliers. One Way ANOVA with Bonferroni post-hoc test, GluA1, df = 52, p = 0.04; α2, df = 52, p = 0.11; α4, df = 52, p = 0.019; α5, df = 52, p = 0.79; α6, df = 52, p = 0.45; β 1, df = 52, p = 0.086; β 2, df = 52, p = 0.48; β 3, df = 52, p = 0.96; δ, df = 52, p = 0.77. Kruskal–Wallis ANOVA on ranks with Dunn's post-hoc test, GluA2, H(1, 55) = 0.48, p = 0.49; GluA3, H(1, 54) = 4.99, p = 0.026; GluA4, H(1, 56) = 3.34, p = 0.068; GluK1, H(1, 56) = 2.6, p = 0.11; GluK2, H(1, 54) = 2.48, p = 0.12; GluK3, H(1, 53) = 1.42, p = 0.23; GluK4, H(1, 53) = 0.0051, p = 0.94; GluK5, H(1, 56) = 7.31, p = 0.0069; GluN1, H(1, 56) = 5.23, p = 0.022; GluN2A, H(1, 56) = 10.28, p = 0.0013; GluN2B, H(1, 57) = 8.35, p = 0.0039; GluN2C, H(1, 55) = 0.19, p = 0.66; GluN2D, H(1, 52) = 5.06, p = 0.025; GluN3A, H(1, 52) = 4.83, p = 0.028; GluD1, H(1, 56) = 7.28, p = 0.007; GluD2, H(1, 53) = 8.33, p = 0.0039; α1, H(1, 49) = 23.62, p = 0.0000; α3, H(1, 50) = 1.47, p = 0.23; ε, H(1, 54) = 0.96, p = 0.33; γ 1, H(1, 53) = 7.03 p = 0.008; γ2, H(1, 58) = 6.31 p = 0.012; γ 3, H(1, 57) = 7.45 p = 0.0063; ρ 2, H(1, 57) = 2.20 p = 0.14; θ, H(1, 50) = 0.85 p = 0.36. *p < 0.05, **p < 0.01, ***p < 0.001.