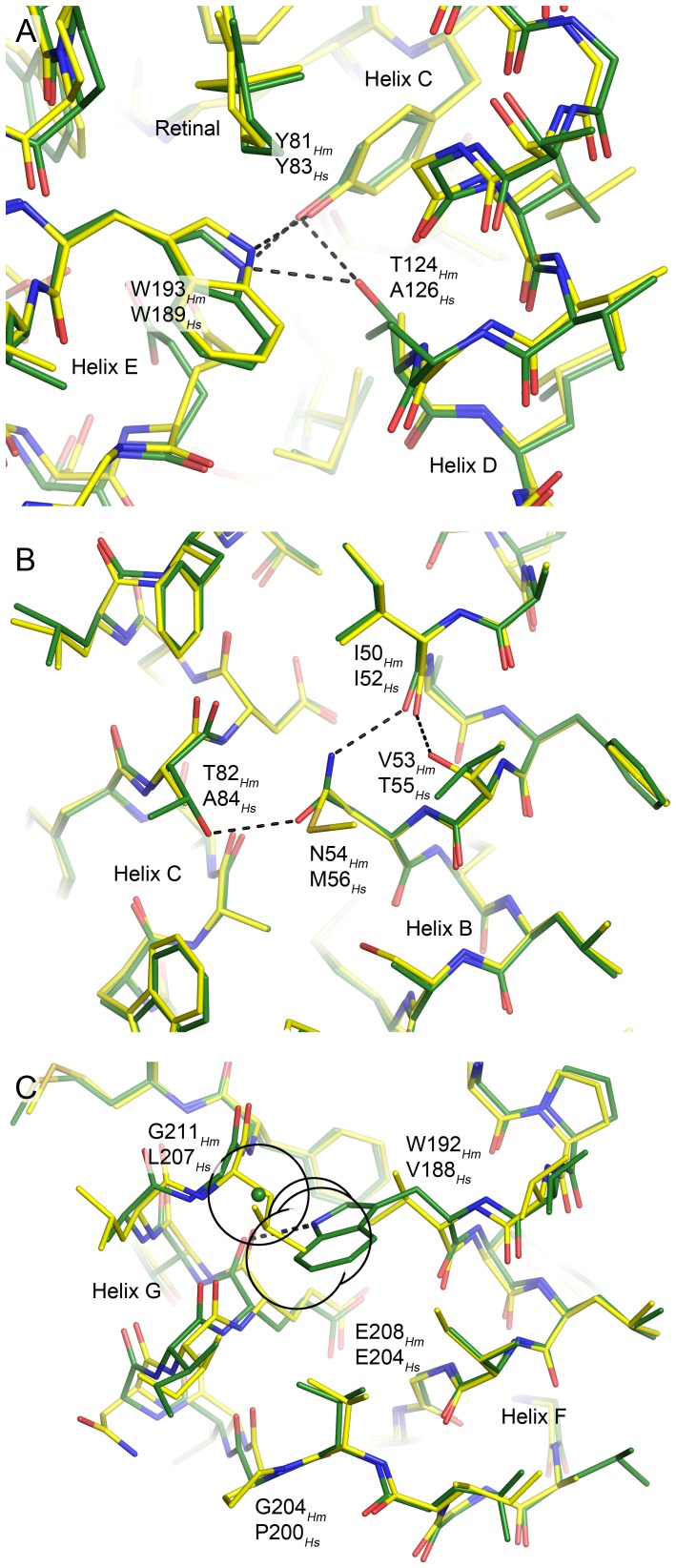
Figure 6. The novel inter-helical hydrogen bonds in HmBRI (green) relative to HsBR (yellow).
In each case, the structure alignment was done locally to emphasize the local effects. (A) The substitution A126Hs → T124Hm results in two novel hydrogen bonds connecting the helix D to helices C and E. (B) The coupled substitution M56Hs → N54Hm, A84Hs → T82Hm results in introduction of the hydrogen bond between the helices B and C. Interestingly, the intra-helical hydrogen bond between the I52 backbone oxygen and T55 is replaced with the hydrogen bond between the homologous I50 backbone oxygen and N54 side-chain amine. (C) Coupled substitution V188Hs → W192Hm, L207Hs → G211Hm results in introduction of the hydrogen bond between the helices F and G (E208 backbone oxygen and W192 indole nitrogen). Interestingly, the glycine is the only possible amino acid at the position 211, as the Cβ atom of any other amino acid would create a steric conflict with W192 side-chain. The hypothetical position of the residue 211 Cβ atom is shown by the green sphere, and its van der Waals radius, as well as those of proximal W192 heavy atoms, is shown as a black circle. Introduction of the bulky tryptophan at the position 192 might be another reason for the divergence of the helices F and G in HmBRI, and, as a consequence, the bigger water-accessible volume of the proton release region.