Figure 2. The live phenotype and positional cloning of the cnot8m1061 mutation.
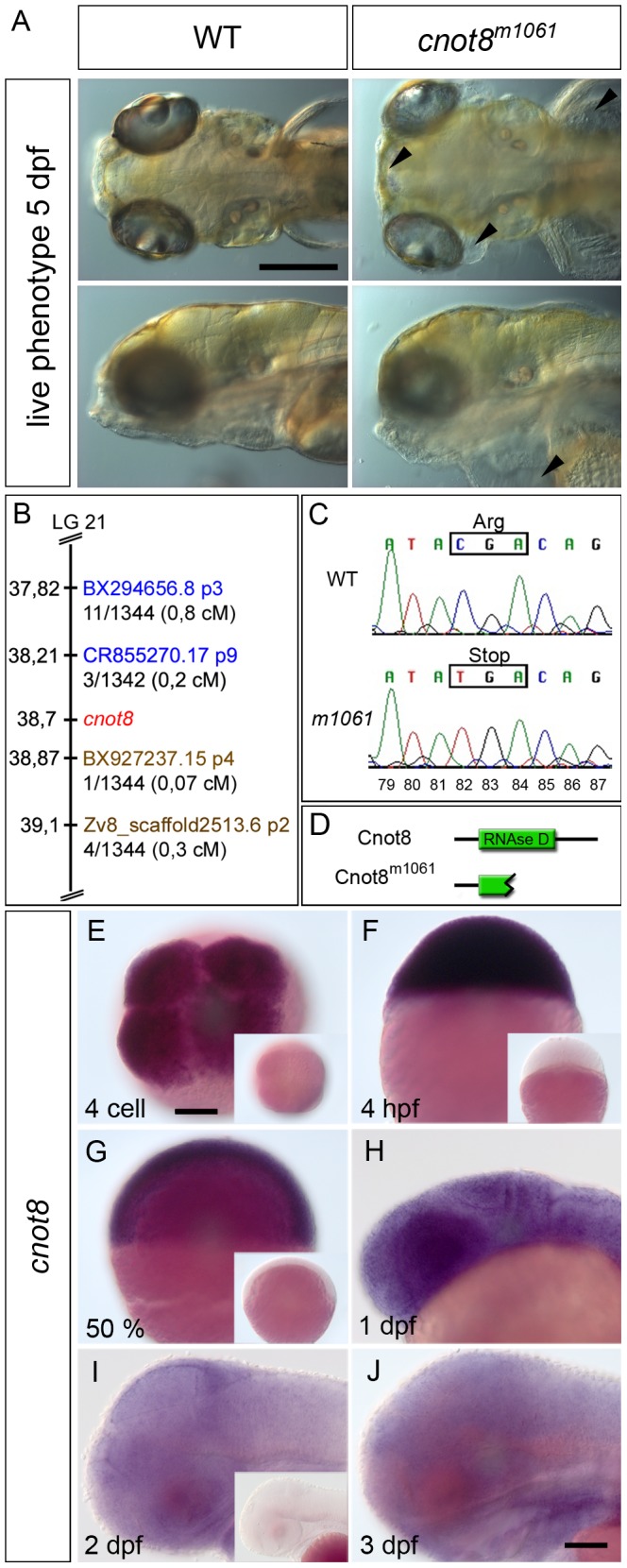
(A) Comparison of cnot8m1061 mutant to wild-type sibling embryos at 5 dpf; cnot8m1061 mutants display edema formation in the eyes and brain (top row, arrowheads) as well as cardiac and yolk sac (bottom row, arrowhead). Top row dorsal view, bottom row lateral view, anterior to the left. Scale bar 100 µm. (B) Scheme of the m1061 genetic interval. Gene and marker positions were obtained from Ensembl Zv8. CR855270.17 p9 and BX927237 p4 define the interval which comprises the genes zgc:77151, MRP L22, gemin5, cnot8, kibra/WWC1, ARL10 and Nop16 (data not shown). Proximal SSLP markers are highlighted in blue. Distal SSLP markers are highlighted in brown. Numbers provided with SSLP markers reflect recombination events identified per number of meioses analyzed. (C) Sequencing of genomic DNA amplified by PCR from individual m1061 homozygous WT and mutant embryos. The C-to-T mutation at bp 82 of the ORF results in the formation of a premature stop codon in m1061 mutant cnot8 ORF. Numbers (79–87) indicate ORF bp position. (D) Zebrafish Cnot8 comprises 286 amino acids and contains an RNAse D domain which is truncated in cnot8m1061 mutants. (E–J) Expression analysis by WISH using cnot8 antisense probes. Sense controls processed exactly in parallel with the antisense reactions are shown as insets in E, F, G and H to evaluate background stain intensities. (E) Maternal mRNA is detected at 4 cell stage (dorsal view). (F) Ubiquitous expression of cnot8 mRNA at 4 hpf (lateral view). (G, H, I) cnot8 continues to be expressed ubiquitously at 50% epiboly, 1, 2 and 3 dpf (lateral views). Scale bars 100 µm.
