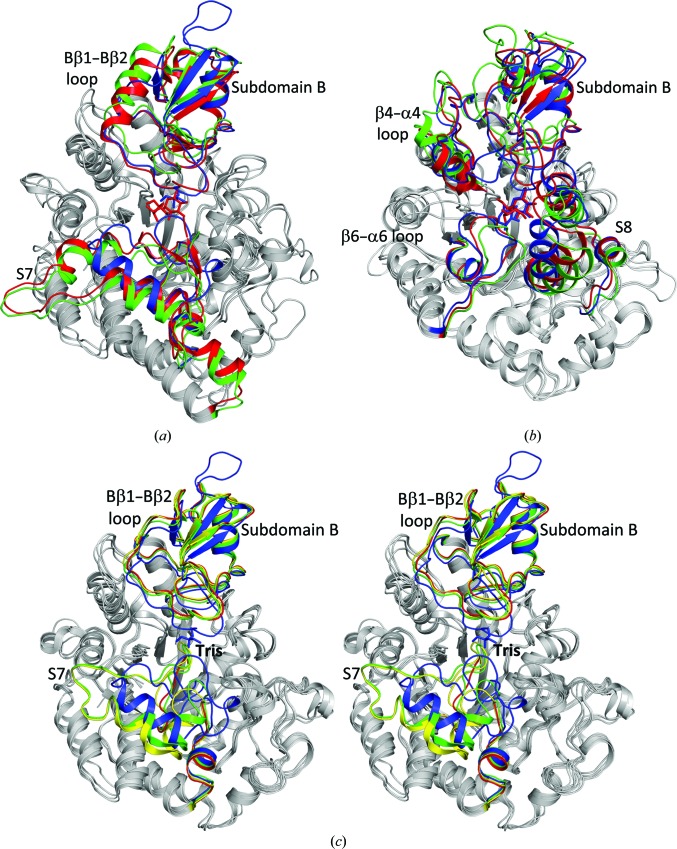
Figure 7.
Different conformations observed in SH, SI and TS. Structural superposition of (a) XaSH-E322Q–sucrose (red), apo XcSH (green; PDB entry 2wpg) and DrTS–Tris (blue) and of (b) RhSI-E254Q–sucrose (red), the RhSI R284C mutant (green; PDB entry 4h2c) and ScIM-E277A–isomaltose (blue; PDB entry 3axh). Some of the member-unique insertions such as subdomains B and S7 in TS, SH and AS, or subdomains B and S and the segments between β4 and α4 and between β6 and α6 in SI and ScIM, modulate the size/shape and accessibility of the active site. These modules may move forward and backward from the active site to form different conformations during enzyme catalysis. (c) Stereoview of the structural superposition of DrTS–Tris, MsTS (PDB entry 3zoa) and MtTS (PDB entry 4lxf) chain A and chain B. The subdomain B and S7 are shaded in blue, green, yellow, and red, respectively. These display distinct tertiary structures of S7 and the relative orientations of the subdomain B to the (β/α)8 barrel are different. A similar rearrangement of subdomain B was observed in TS, AS and SH.