Table 1. Statistics of data collection and structural refinement.
Values in parentheses are for the highest resolution shell.
| Wild type | N253A | |
|---|---|---|
| PDB code | 4tvu | 4wf7 |
| Data collection | ||
| Space group | P21 | P212121 |
| Unit-cell parameters (, ) | a = 95.61, b = 195.90, c = 130.99, = = 90, = 92.89 | a = 96.25, b = 133.71, c = 196.61, = = = 90 |
| Resolution () | 302.70 (2.802.70) | 202.21 (2.292.21) |
| Observed reflections | 462831 (38844) | 759037 (71092) |
| Unique reflections | 128557 (11771) | 129348 (12695) |
| Completeness (%) | 97.7 (89.4) | 99.9 (99.7) |
| Multiplicity | 3.6 (3.3) | 5.9 (5.6) |
| I/(I) | 13.1 (2.9) | 12.6 (2.3) |
| R merge † (%) | 9.6 (40.8) | 15.1 (78.7) |
| Refinement | ||
| Resolution () | 302.7 (2.772.70) | 202.21 (2.272.21) |
| Reflections [F> 0(F)] | 122065 (8113) | 119863 (8156) |
| R cryst ‡ (%) | 19.4 (28.2) | 16.0 (22.6) |
| R free § (%) | 23.6 (31.9) | 20.0 (26.3) |
| R.m.s. deviations | ||
| Bond lengths () | 0.01 | 0.01 |
| Bond angles () | 1.42 | 1.41 |
| Mean B value (2) | ||
| Protein | 38.0 | 33.0 |
| Metal ions | 27.1 | 26.7 |
| Tris | 32.5 | 35.9 |
| Water | 24.9 | 37.3 |
| Ramachandran analysis¶ (%) | ||
| Favoured | 97.4 | 97.4 |
| Allowed | 2.6 | 2.6 |
| Disallowed | 0 | 0 |
R
merge = 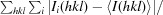
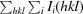 , where Ii(hkl) is the average intensity value of the equivalent reflections.
, where Ii(hkl) is the average intensity value of the equivalent reflections.
R
cryst = 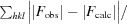
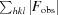 , where F
obs and F
calc are the observed and the calculated structure factors, respectively.
, where F
obs and F
calc are the observed and the calculated structure factors, respectively.
R free was calculated using 5% of data that were randomly excluded from refinement.
The Ramachandran analysis was performed by MolProbity (Chen et al., 2010 ▶).
