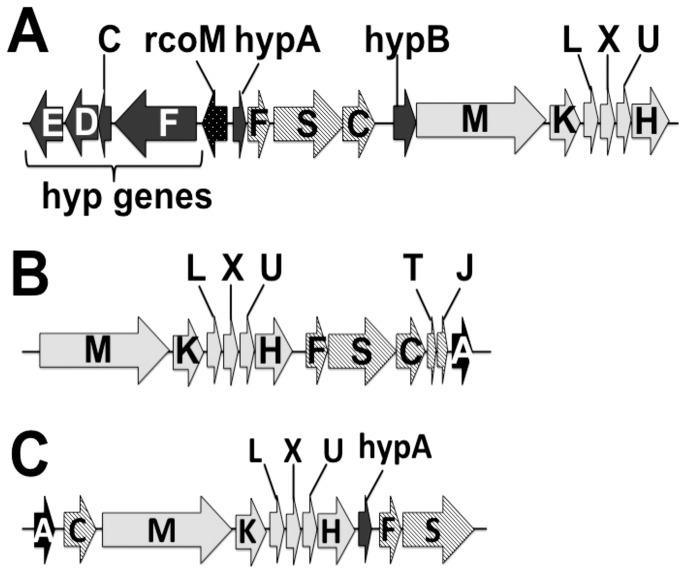
Figure 3. Organization of genes encoding anaerobic CO metabolism in multiple bacteria.
(A) Gene structure of CO metabolism genes in Rubrivivax gelatinosus CBS. The hyp genes putatively responsible for coo hydrogenase maturation are clustered among the coo CODH and hydrogenase genes. The gene encoding RcoM, the putative transcription factor for the coo CODH and hydrogenase genes is located among the hyp genes within this cluster of CO metabolism genes. (B) Gene structure of CO metabolism genes in Rhodospirillum rubrum. The cooA gene, encoding the CooA CO-responsive transcription factor, directly follows the CODH genes. (C) Gene structure of CO metabolism genes in Carboxydothermus hydrogenoformans. C. hydrogenoformans has five CODH complexes, but the genes for only one CODH are co-located with the genes for an energy-conserving hydrogenase [36]. The cooA and hypC genes precede the cooMKLXUH hydrogenase genes, which are followed by hypA and the cooFS CODH genes.