Abstract
A pilot-scale multilayer rapid infiltration system (MRIS) for domestic wastewater treatment was established and efficient removal of ammonia and chemical oxygen demand (COD) was achieved in this study. The microbial community composition and abundance of ammonia oxidizers were investigated. Efficient biofilms of ammonia oxidizers in the stationary phase (packing material) was formed successfully in the MRIS without special inoculation. DGGE and phylogenetic analyses revealed that proteobacteria dominated in the MRIS. Relative abundance of ammonia-oxidizing archaea (AOA) and ammonia-oxidizing bacteria (AOB) showed contrary tendency. In the flowing phase (water effluent), AOA diversity was significantly correlated with the concentration of dissolve oxygen (DO), NO3-N and NH3-N. AOB abundance was significantly correlated with the concentration of DO and chemical oxygen demand (COD). NH3-N and COD were identified as the key factors to shape AOB community structure, while no variable significantly correlated with that of AOA. AOA might play an important role in the MRIS. This study could reveal key environmental factors affecting the community composition and abundance of ammonia oxidizers in the MRIS.
Introduction
Nowadays human activities harmfully affect limited freshwater resources. Freshwater resources on Earth are diminishing rapidly, making water resource conservation and regeneration a serious challenge for human beings. Efficient water reuse techniques play an important role in wastewater treatment. Intermittent infiltration systems are among the most promising systems due to their simplicity, reliability, low energy consumption and low cost. Such systems combine the complex effect of physical filtration, chemical reaction and biological transformation, thus can achieve high purification efficiency for domestic wastewater treatment. The biological transformation mainly refers to microbial nitrification and denitrification, is gaining more and more attention recently due to its significant contribution to the nitrogen removal in these systems [1].
Nitrification is the microbial oxidation of ammonia to nitrate via nitrite as intermediate, which plays an important role in the global nitrogen cycle and in controlling effluent toxicity in wastewater treatment [1]. Ammonia-oxidizing microorganisms (AOM), including ammonia-oxidizing archaea (AOA) and ammonia-oxidizing bacteria (AOB), are thought to be the main ammonia oxidizers. Both AOA and AOB contain ammonia monooxygenase (AMO) which catalyzes the first rate-limiting step of ammonia to hydroxylamine [2]. Autotrophic AOB including Nitrosomonas communis, Nitrosococcus mobilis, and N. halophilus affiliate with β- and γ-proteobacteria, had been considered to be the most important contributor to ammonia oxidation for a long time [3]. However, recent investigation found that AOA would also be responsible for nitrification. At present, thermophilic strains of AOA had been cultivated, such as Candidatus Nitrosocaldus yellowstonii and Candidatus Nitrososphaera gargensis, implying a broad distribution of ammonia oxidation among crenarchaeota, which reinvigorates the debate on the thermophilic ancestry of AOA [4], [5]. In certain environmental conditions, AOA even contribute more to microbial nitrification than AOB. Numerical dominance of archaeal over bacterial ammonia oxidizers in soil ecosystems indicated that crenarchaeota migh51ht be the most abundant AOM [6]. Exclusive growth of archaeal ammonia oxidizers revealed that ammonia oxidation under active nitrification condition was mainly due to AOA nitrification [5], [7].
Present reports showed that in wastewater treatment system (WWTS) physiological and ecological difference occurred among differences AOM genera and lineages in response to different environmental factors such as substrate concentration, temperature, salinity, pH, biogeography, and so on [8]–[10]. It can be expected that the AOM community structure also changes in the pilot-scale rapid infiltration system (MRIS) established in this article in response to different treatments. Hence, investigating the community structure of ammonia oxidizers in the MRIS induced by these environmental factors will improve our understanding about their roles in the nitrogen cycling in terrestrial ecosystems, and supply feedback to reevaluate WWTS. Present methods/techniques for investigation of AOM community structure consist of culture methods and biochemical techniques, etc. However, all culture methods are potentially selective and thus bear the risk of incomplete coverage of the actually existing bacterial diversity [11]. Furthermore, most AOM is difficult to cultivate in laboratory, which prevents its natural community structure analysis. Uncultured techniques based on in situ detection of 16S rRNA and amoA genes such as denaturing gradient gel electrophoresis (DGGE) can supply useful information about the microbial diversity in comparison to laboratory culture methods [12]. Moreover, quantitative analysis of the nitrifying indicator amoA genes by real-time quantitative polymerase chain reaction (qPCR) allows fast, sensitive, and simple inspection of variation of AOM abundance in WWTS [3].
In this study, we established a modified intermittent infiltration system named MRIS and evaluated its contaminant removal efficiency with several criteria including chemical oxygen demand (COD), NH3-N, NO3-N, pH, dissolved oxygen (DO), and total nitrogen (TN). Moreover, we analyzed the microbial community structure using DGGE profiles based on 16S rRNA gene and amoA gene sequences. Finally, we quantitated the amoA genes of different samples collected from different spots of the MRIS by qPCR technique, with the purpose of elaborating the correlation between the criteria detected and the microbial community.
Materials and Methods
Construction of the pilot-scale MRIS
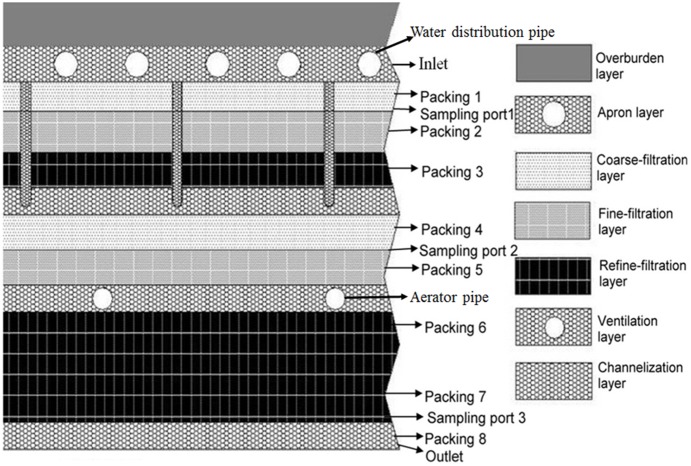
The MRIS was established outdoor and was operated nearby an apartment in the yard of Guangzhou Institute of Geochemistry (the field study did not involve endangered or protected species; location: latitude/longitude: 23.129163N/113.264435E; and no specific permissions were required for this location). Main part of the system is the filter sheet, which was stacked with sands of different diameters to form 11 layers in a leak proof cement pond. The total height of the filter sheet is 100 cm and length/width is 100 cm/100 cm. The crucial structure layers include the coarse-filtration layer (effective size d10 = 0.19, d60 = 0.61, uniformity coefficient = d60/d10 = 3.2), fine-filtration layer (effective size d10 = 0.17, d60 = 0.62, uniformity coefficient = d60/d10 = 3.6) and the refine-filtration layer (effective size d10 = 0.15, d60 = 0.62, uniformity coefficient = d60/d10 = 4.1). Thickness of packing 1 is 6 cm, packing 2 is 8 cm, packing 3 is 7 cm, packing 4 is 8 cm, packing 5 is 8 cm, total thickness of packing 6 and 7 is 25 cm, and packing 8 is 6 cm. Water distribution pipes were placed in the Apron layer and aerator pipes were placed in the Ventilation layer (Figure 1). The MRIS was fed with domestic wastewater but not fecal sewage, and was automatically operated. In order to prevent choking, wastewater was poured into a sedimentation tank via water distribution pipe before being pumped in to the filter system. Wastewater was intermittently pumped from the sedimentation tank into the filter, the pump was automatically turned on eight times per day and was operated for 27 min each time, with a total loading rate of 0.5 t/m2·d. The detail running times were 0∶00–0∶27, 3∶00–3∶27, 6∶00–6∶27, 9∶00–9∶27, 12∶00–12∶27, 15∶00–15∶27, 18∶00–18∶27, 21∶00–21∶27. The MRIS was aerated intermittently twice during the hydraulic retention time, 10 minutes each time through the ventilation layer (e.g. between 0∶27 to 3∶00, the MRIS was aerated in 1∶30–1∶40 and 2∶30–2∶40) (Figure 1).
Figure 1. Structure of the filter sheet and sampling ports.
Sampling and chemical analysis
According to pre-experiments, the treatment efficiency of the MRIS reached steady state after being operated for 4 months. Four water sampling ports (Sampling port 1–3 and the Outlet) were set in different layers in the filter sheet (Figure 1). As microbial biomass varied considerably in the filter sheet, different volumes of water sample in different sampling ports were sampled to obtain approximately biomass. 300 mL raw wastewater, 500 mL, 1000 mL, 3000 mL, 5000 mL treated wastewater effluent from the Inlet (assigned as A1, similarly hereinafter), Sampling port 1 to 3 (sequentially A2–4), and the Outlet (A5) were collected in order (Figure 1). Triplicate water samples were collected eight times on the same day and were respectively mixed as one sample. Packing material was collected in different depths of the filter sheet, i.e., Packing 1 to 8 (assigned as P1–8 sequentially) (Figure 1). Water content of the packing material was measured within 24 h.
NH3-N was measured colorimetrically according to the Standard Methods [13]. NO3-N and NO2-N were determined with an iron chromatography (Shimadzu SCL-10ASP, Japan). The pH value was determined potentiometrically using a pH analyzer (Sartorius PB-20, Germany). The DO level was analyzed with a digital, portable DO meter (HQ30d, America).
DNA extraction and PCR amplification
For DNA analysis, water samples were filtered respectively over 0.22 µm pore size polycarbonate membranes (45 cm×diameter), the filter cake were stored at −30°C until use. In order to get rid of big particles, all water samples were centrifuged at 500 rpm for 15 min at room temperature. The packing material was first washed by ultrapure water to remove humus, and then was shocked with glass bead to obtain microbial biomass. Genomic DNA was extracted from filter cake (about 0.2 g) or washed packing material (about 0.5 g) by UltraClean Soil DNA Kit (MoBio Laboratories, Solana Beach, CA, USA).
Nested PCR was conducted to increase the sensitivity of DGGE profile of 16S rRNA gene. In the first round, 27F/1492R primers were used according Brosius et al [14]. During the second round, 16S rDNA fragments were reamplified using bacterial primers 338F/534R [15]. For DGGE profile of amoA gene, bacterial amoA gene specific primers amoA-1F-26 GC (containing GC clamp) and amoA-2R [16], archaeal amoA gene specific primers arch-23F/arch-616R (both without GC clamp) were used [17]. The PCR mixture was prepared with 5 µL of 10×PCR buffer, 2 µL of dNTPs mixture (TaKaRa, China), 1 µL of each primer (10 µM), 1 µL of DNA extract, 0.5 µL of Taq polymerase, 1 µL of bovine serum albumin (BSA, 0.1%), and was adjusted to a final volume of 50 µL with ddH2O.
DGGE profile of 16S rRNA and amoA genes
DGGE profile was carried out in 8% (w/v) polyacrylamide gels with a denaturing gradient of 35–75%, 0–45% and 40–70% for 16S rDNA V3 region PCR products, archaeal and bacterial amoA gene PCR products, respectively. Electrophoresis was performed in 1×TAE buffer at 60°C, 80 V for 12.5 h. Gels were stained for 20 min in 150 mL 1×TAE buffer containing 100 ng/mL Goldviewer dye. Visualization and digital photography was acquired with a CCD camera controlled by Quantity One software (Bio-Rad, USA) [16]. Major bands were excised and reamplified, PCR products were cloned into pMD19-T Simple Vector (TaKaRa, China). Four to six randomly selected clones containing correct insert size from each DGGE band were sequenced using M13–47 primer. Positive sequences were aligned using BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi) and Clustal X 1.83. A phylogenetic tree was constructed using the Neighbor-Joining method with Jukes-Cantor correction by MEGA v4.0.2 software [18]. Robustness of tree topology was verified by calculating bootstrap values of 1000 replications for the Neighbor-Joining tree as previously described [19].
qPCR analysis for AOB and AOA amoA gene
The abundance of AOB and AOA amoA genes was quantified with qPCR using previously described primers (amoA-1F and amoA-2R for AOB, amo196F and amo277R for AOA) [19], [20]. The reaction mixture contained 25 µL of 2×IQTM SYBR green supermix (TaKaRa, China), 10 pmol of each primer, and 20 µg BSA in a final volume of 50 µL. Amplification, detection, and data analysis were performed in triplicate using the Eppendorf MasterCycler ep Realplx4 system (Eppendorf, Germany). A control was always run with water as template instead of AOB or AOA DNA extract. Specific amplification of AOB or AOA amoA genes were confirmed by melting curve analysis always resulting in a single peak and by agarose gel electrophoresis. The fluorescence signal of the amplified DNA was used to quantify the concentration of AOB and AOA amoA genes [21]. Quantification was based on the comparison of the C T value between samples and the calibration curve of amoA gene standard. The AOB and AOA numbers were calculated by assuming two or one amoA gene copy number(s) per cell, respectively [20], [22].
Statistical analysis
SPSS 16.0 was used to evaluate the correspondence between the Shannon diversity index, environmental variables and amoA gene abundance. Bivariate followed by Pearson and two-tailed was used to check for the correlation coefficients, p<0.05 was considered to be statistically significant and p<0.01 was highly significant. To investigate the correlation between microbial community composition and environmental variables, CANOCO for windows (version 4.5) was used for multivariate statistical analysis. The relative intensity of each DGGE band represented microbial community composition of effluent. Detrended correspondence analyses (DCA) of DGGE band matrices indicated that the maximum of lengths of gradient was below 3, so redundancy analysis (RDA) was performed [23]. The significance of the relationship of environmental variables to the variation in microbial community composition was tested using Monte Carlo tests (999 unrestricted permutations, p<0.05).
Nucleotide sequence accession numbers
The sequences reported in this study have been deposited in GenBank under the accession numbers of JQ963286 to JQ963324 for bacterial 16S rDNA V3 region, and KF460097 to KF460108 for amoA genes of AOB and AOA.
Results
Treatment performance of the MRIS
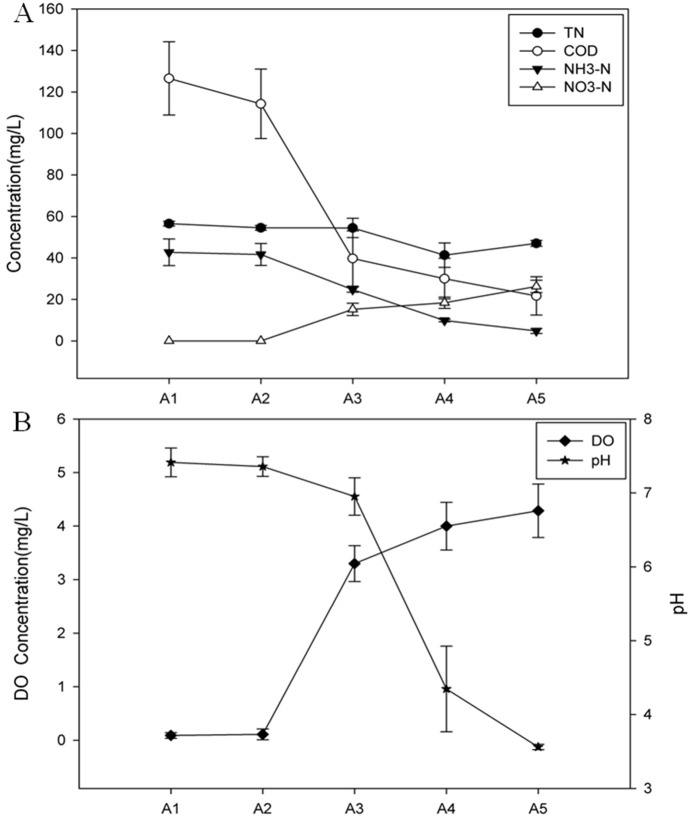
The constructed MRIS exhibited excellent performance without special inoculum. The loading burden could reach to 0.5 t/m2·d. As the system had been in steady state after running for 4 months, water quality could be regarded to be at the homogeneous level. To further assure the stability and representativeness of the analyses, water samples used were collected in triplicate and mixed as one final sample for all experiments, as indicated by the Sampling and chemical analysis above (see Materials and Methods). The structure of Ammonia-oxidizing microorganism community can be significantly affected by temperature in an infiltration bioreactor, and the DGGE fingerprints of AOM communities will consequently change according to the ambient temperature. However, in this study, the temperature of all samples could be treated as the same, as triplicate water samples were moderately and intermittently collected eight times on the same day and were respectively mixed as one sample. The pH value decreased from 7.41 to 3.56 along the depth of the filter sheet, which suggested that acidifying nitrification process occurred (Figure 2B). DO was only 0.9 mg/L in the upper layer of the filter sheet, while in the Outlet (A5) it increased dramatically to 4.29 mg/L (Figure 2B). The total removal ratio of COD and NH3-N in the MRIS were 82.8% and 88.7%, respectively (Figure. 2A). Between sampling port 1 (A2) and 2 (A3), COD removal ratios reached the highest (Figure 2A) and DO increased dramatically (Figure 2B). No or very low nitrite could be detected in raw wastewater (A1) and the ongoing-processed wastewater sampled from A2, while in the following sampling ports (A3 and A4) and the Outlet (A5), nitrate concentration increased evidently along the depth of the filter sheet (Figure 2A). All these results indicated that efficient microbial nitrification was achieved in this MRIS.
Figure 2. Performance of the MRIS for domestic wastewater treatment.
(A) The concentration of chemical oxygen demand (COD), total nitrogen (TN), ammonia nitrogen (NH3-N), nitrate nitrogen (NO3-N); (B) Dissolved oxygen (DO) concentration (the right axis) and pH value (the left axis) in the Inlet, Sampling ports and the Outlet. A1–5: the Inlet, Sampling port 1–3, and the Outlet, correspondingly.
Community diversity and distribution of AOB based on 16S rRNA gene
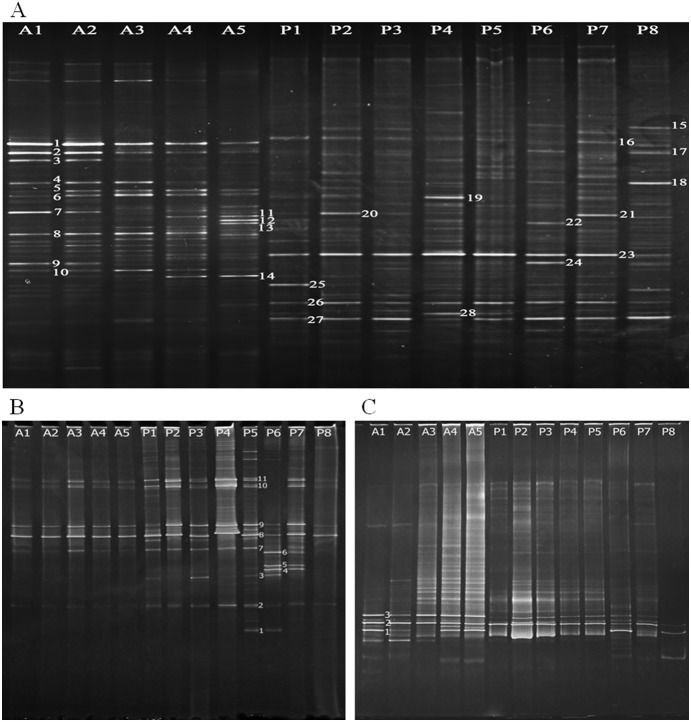
Major DGGE bands of bacterial 16S rRNA gene were re-amplified and evaluated phylogenetically in order to identify putative functional bacteria in the flowing (water effluent) and the stationary phase (packing material) of the MRIS. DGGE profile of bacterial 16S rRNA gene showed frequent shifts in the composition and diversity of microbial community, as indicated by the appearance and disappearance of certain bands in different samples obtained from different sampling spots (Figure 3A). 28 bands were recovered and used for phylogenetic analysis (Figure 4). The relative abundance of the 16S rRNA genes indicated the richness of AOB in the MRIS. The Shannon index revealed that the community diversity between the flowing and the stationary phase did not vary significantly.
Figure 3. DGGE profiles of 16S rRNA and amoA genes.
16S rRNA genes (A), AOB amoA genes (B), and AOA amoA genes (C) were amplified from the flowing (A1–5) and the stationary phase (packing material) (P1–8), respectively. Electrophoresis was carried out at a constant voltage of 80 V at 60°C for 12 h. Gels were stained with GoldViewer dye. A1–5: the Inlet, Sampling port 1–3, and the Outlet, correspondingly; P1–8: Packing 1–8, correspondingly.
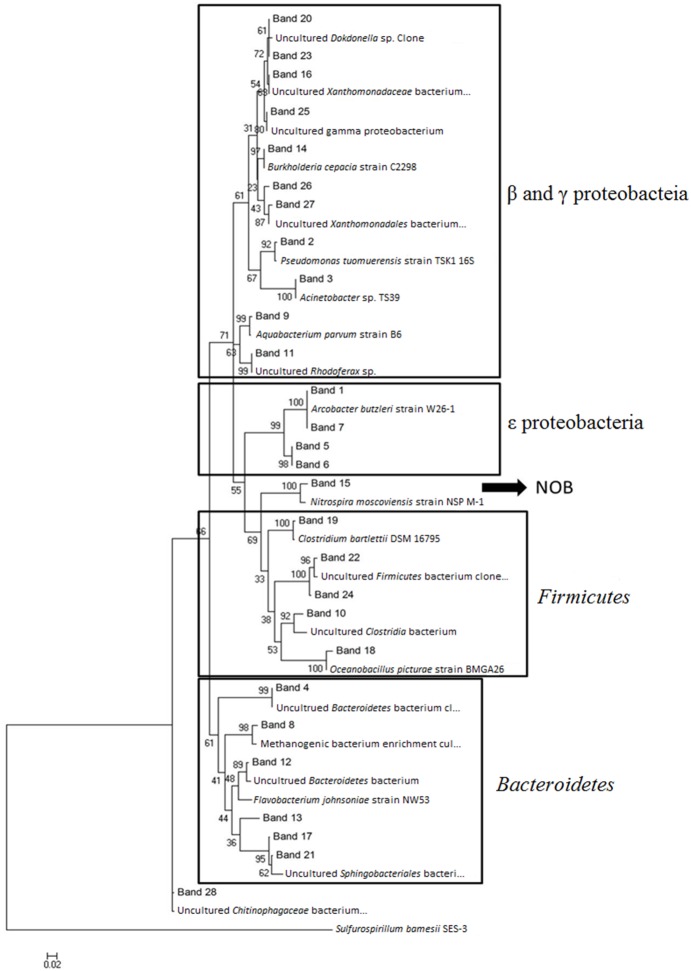
Figure 4. Phylogenetic analysis of 16S rRNA genes.
Phylogenetic tree was constructed using the Neighbor-Joining method by MEGA v4.0.2 software. The numbers at the nodes are bootstrap values (n = 1000) and the Random seed value is 64,238.
Phylogenetic analysis based on the retrieved 16S rRNA gene sequences showed generally four clusters (Figure 4). Three major groups affiliated to the phyla of proteobacteria (53.6%), firmicutes (17.9%), and bacteroidetes (21.4%), correspondingly. As in many WWTS, proteobacteria dominated the microbial population in the MRIS, among which β-, γ-, and ε-proteobacteria accounted for 10.7, 28.6, and 14.3% of the total clones, respectively. Clostridia sp. was simultaneously detected both in the flowing (band 10) and stationary phase (band 19). In addition, one band (15) was found to be directly relative to nitrification, which shared up to 97% sequence similarity to Nitrospira moscoviensis. Five bands (10, 18, 19, 22, and 24) exhibited the closest phylogenetic affinity to the firmicutes phylum. ε-proteobacteria were also detected and were clustered together: band 1 and 7 shared 100% sequence similarity to Arcobacter butzleri; band 5 and 6 shared 100% sequence similarity to Sulfurospirillum barnesii. The community diversity of AOB in the MRIS seems to be relatively low based on the DGGE profile of 16S rRNA gene.
Analysis of community structure and composition of AOM based on amoA gene
Compared with 16S rRNA genes, detection of the amoA genes encoding the subunit of AMO could be more sensitive and specific for the analysis of community diversity and functional AOM abundance in the MRIS. Therefore, DGGE analysis based on specific ammonia oxidizing indicator amoA gene was conducted to assess the AOM community structure and composition. DGGE profiles based on AOB amoA gene exhibited nearly identical feature in all samples from the flowing phase, as shown by Figure 3B. The Shannon index showed that the AOB community of the stationary phase was more diverse than that of the flowing phase. Nine bands were selected for further BLAST and phylogenetic analysis (Figure 4), which revealed that 1) most of the closest relative sequences came from uncultured AOB; 2) the wastewater treatment plants rarely harbored Nitrosospira [24], nevertheless, bands obtained here exhibited great sequence similarity to Nitrosospira sp., band 1, 2 and 5 possessed up to 95, 99 and 96% sequence similarity to uncultured Nitrosospira sp. PJA1 (GenBank: DQ228457.1), N. multiformis ATCC 25196 (GenBank: DQ228454.1) and Nitrosospira sp. analogues (GenBank: GU136449.1), correspondingly, and band 3 showed 99% sequence similarity to Nitrosolobus multiformis analogue (GenBank: U91603.1); 3) all retrieved bands affiliated to β-proteobacterium (Table S1).
Three bands from DGGE profile of AOA amoA gene were successfully excised and sequenced (Figure 3C). BLAST analysis revealed that all the closest relative sequences deposited in GenBank came from moderately thermophilic AOA Crenarchaeote. Specifically, band 2 shared 99% sequence similarity to AOA discovered in acidic soil. In comparison with the relative stability of AOB community of the flowing phase, the AOA community of the flowing phase showed an increasing Shannon index along the depth of the filter sheet. However, the AOA diversity was relatively lower than the AOB diversity in the stationary phase (Figure 3B and Table S2).
AOB and AOA abundance in the MRIS
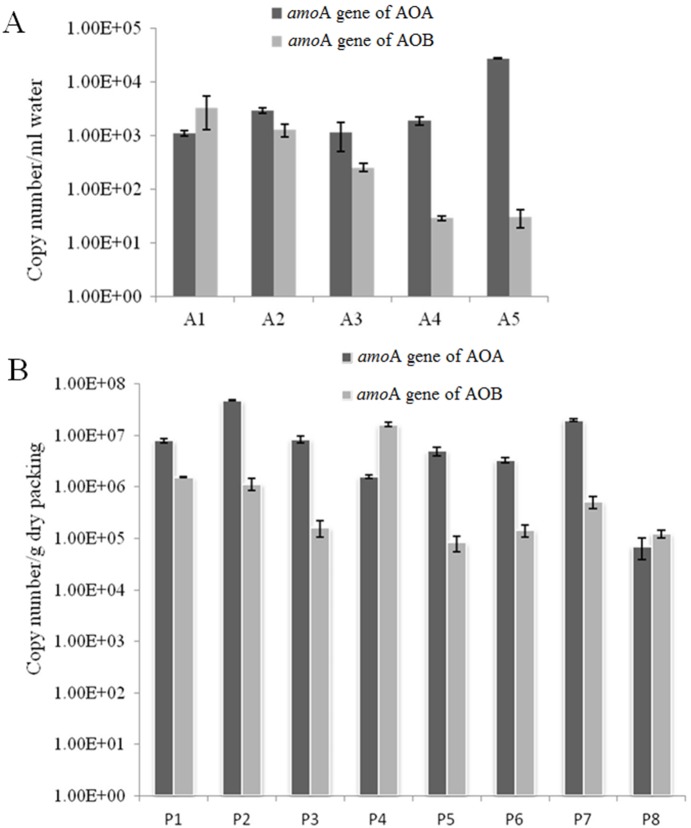
Genetic diversity of AOB and AOA amoA genes from different samples was analyzed by qPCR. In the flowing phase, AOA abundance generally showed an imperfect increasing trend: it was relatively low in the Inlet (A1), but reached its peak in the Outlet (A5) (Figure 5A). The copy numbers of AOA amoA gene ranged from 1.11×103 copies/mL to 2.75×104 copies/mL (Figure 5A). However, AOB abundance showed an unambiguous decreasing trend along the depth of the filter sheet (Figure 5A). The copy numbers of AOB amoA gene ranged from 3.33×103 copies/mL in the Inlet (A1) to 3.04×101 copies/mL in the Outlet (A5). Furthermore, the abundance of AOA was greater than that of AOB in the flowing phase. However, the copy numbers of AOM amoA genes in the stationary phase fluctuated, no clear trend was observed. The copy numbers of AOA amoA genes ranged from 7.05×104 to 4.82×107 copies per gram dry weight, and that of AOB amoA genes from 8.31×104 to 1.63×107 copies per gram dry weight (Figure 5B). Neither AOA nor AOB abundance was significantly decrease or increase along with the depth of the filter sheet, and thus inconspicuous trend of AOB and AOA abundance was observed in the stationary phase (Figure 5B). Though the pH and ammonia decreased constantly (Figure 2), inconsistent trend of AOB copies displayed in the packing matrix was observed (Figure 5B). Such observation might be attributed to the different sizes of sands in the MIRS; moreover, the living bacteria proportion in the biofilm of the packing samples is hard to estimate.
Figure 5. The copy numbers of AOB and AOA amoA genes from the flowing (A) and the stationary phase (B).
All data are the means of values obtained from three parallel experiments ± SD (t-test, p<0.01) using the ΔΔCT method. A1–5: the Inlet, Sampling port 1–3, and the Outlet, correspondingly; P1–8: Packing 1–8, correspondingly.
Effects of environmental factors on AOM communities
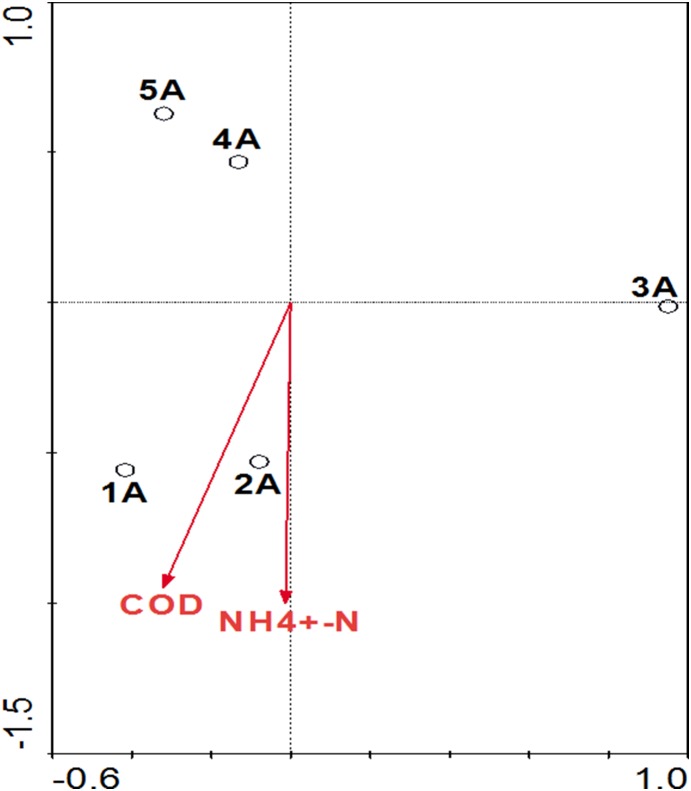
The relationship of the Shannon diversity index, amoA gene abundance and environmental variables was evaluated by SPSS 16.0. In the flowing phase, there was a significant positive correlation between DO concentration and AOA Shannon index (r = 0.963, p<0.05). Meanwhile, NH3-N showed a highly significant negative correlation (r = −0.998, p<0.01), whereas NO3-N showed a significant positive correlation (r = 0.965, p<0.05) with AOA Shannon index. No significant correlation was observed between AOB Shannon index and chemical indices in this study. Despite the fact that AOB abundance significantly correlated with DO concentration negatively (r = −0.998, p<0.01), significant positive correlation between AOB abundance and COD concentration (r = 0.997, p<0.01) was observed. However, there was no significant correlation between AOA community structure and environmental variables. When both COD and NH3-N were treated as environmental variables, the cumulative percent variance of the species-environment relationship indicated that the first and second canonical axes accounted for 76.8% and 6.8%, respectively. Moreover, Monte Carlo tests for the first axis and all canonical axes were significant (p<0.05) (Figure 6).
Figure 6. Triplot of RDA for AOB in the flowing phase.
The first and the second axes explained 76.8% and 6.8% of the total variation respectively. The length of each arrow is correlated with the degree of relationship between the response variables. The arrows point in the direction of the maximum change for the associated variable. Open symbols represent samples from the flowing phase.
Discussion
Effective ammonia removal in wastewater is mainly due to the combined effect of AOB and AOA. Certain environmental factors can affect both AOB and AOA abundance, such as ammonia, organic carbon, temperature, salinity, DO level, pH, nitrogen and carbon sources, sulfide, and phosphate, and so on [10], [24]. Among these factors, the ammonia level, salt concentration, DO density and pH value are the most important factors.
Microbial nitrification is a proton releasing reaction, which oxidizes ammonia to nitrite (NH4 + +2O2 → 2H+ + NO3 − + H2O). Accordingly, pH value along the depth of the filter sheet decreased as nitrification proceeded (Figure 2B). It was shown that nitrification could be down-regulated or even completely inhibited when the microenvironment pH was under 6.45 or above 8.95 [25]; nitrification in biofilms grown on chalk particles reached its peak at a pH around 4 owning to nitrifying populations related to subgroups with low Kms for ammonia [26]. However, it seemed that the oxidization of ammonia and nitrite in the MRIS maintained under extremely acidic condition (Figure 2). Evidence revealed that AOA were more predominant than AOB in acidic soils, where inhibition of AOB growth under acidic condition was observed [27]. AOB were sensitive to pH and could only thrive at neutral to alkali environments [24], but AOA were more tolerant to low pH. It appeared that AOA had a wide ecological and phylogenetic diversity under a broad range of pH [10]. For these reasons, a gradual decrease of AOB amoA gene abundance along the depth of the filter sheet could be expected in the MRIS, with an overall increase of AOA amoA gene abundance observed in the flowing phase (Figure 5A), where reducing ammonia and pH were monitored (Figure 2A and B). These results implied that AOA contributed more in the MRIS than AOB did.
It’s known that DO could suppress AOB activity at low concentration, resulting in limited nitrification [25] and leading to high COD [28]. Therefore, maximum AOB abundance (Figure 5A) in the superficial layers of the MRIS, where very low DO and high COD concentration were observed, might be attributed to the low DO concentration in this system (Figure 2A and B). Though considerable biomass, maximum AOB abundance and comparable AOA, was detected, NO3-N was undetectable because nitrification did not start up or was weak at the early stage in the superficial layers (Figure 2A). High DO led to oxidization of nitrite to nitrate and inefficient denitrification [29], causing nitrate accumulation in the Outlet (A5). Therefore, suppression of AOB with increasing nitrate was observed, when DO increase along the depth of the filter sheet (Figure 2A).
AOA community diversity in the MRIS (Figure 3C) positively correlated with DO concentration as indicated by Shannon index (r = 0.963, p<0.05), Meanwhile, NH3-N displayed a significantly negative correlation (r = −0.998, p<0.01) with DO concentration. These results implied that DO and NH3-N were two important factors for shaping AOA community. The changes of AOA community structure in the flowing phase also demonstrated that the influent wastewater fed the packing (stationary phase) to form biofilms and led to partial cell accumulation. Though no significant correlation was observed between AOB Shannon index and chemical indices, a close association of RDA indicated that COD and NH3-N were best correlated with the AOB distribution in the MRIS (Figure 6), as RDA showed that the selected parameters were fine explanatory variables of the AOB community structure.
Ammonia has two opposite effects on the growth of AOB. High ammonia promotes AOB growth, while low ammonia inhabits its growth [30]. In contrast to AOB, AOA can endure very low ammonia concentration [31]. Along the depth of the filter sheet, the growth of AOB was suppressed but that of AOA was strengthened (Figure 5A) when ammonia concentration decreased (Figure 2A). In soil samples, AOB and AOA also preferred different N conditions for growth: AOB required high ammonia, while AOA required low ammonia [32]. Similar case in this study revealed different contribution of AOB and AOA to N cycling in the MRIS, AOA may play more important role in ammonia oxidization in the flowing phase. When DO level was taken into consideration, confusion arose to some extent due to the reason that AOA were generally thought to be more active in low-oxygen and oxic-anoxic surroundings [33], while most AOB preferred aerobic condition [34]. Thought significant negative correlation between AOB abundance and DO concentration (r = −0.998, p<0.01) and significant positive correlation between AOB abundance and COD concentration (r = 0.997, p<0.01) were observed, DO level seemed to be a relative “trivial” factor for AOA in the MRIS, as both DO and COD concentration had no significant correlation with AOA abundance. Yet, fully aerobic growth of Nitrosopumilis maritimus during cultivation and near-stoichiometric conversion of ammonium to nitrite were reported [35]. Thus we could not rule out the possibility that some aerobic AOA might exist in the MRIS and contribute to nitrification in spite of ‘toxic’ oxygen. The population feature in the stationary phase was characterized by divergent expression level of amoA genes (Figure 5B). In combination with AOM abundance in the flowing phase, the copy numbers of AOA amoA genes exceeded those of AOB in the MRIS, as microbial nitrification went on along the depth of the filter sheet.
Phylogenetic analysis based on 16S rRNA gene sequences showed relative similarity to the rotating activated bacillus contactor (RABC) [36], where three major groups were found (Figure 4). Among the firmicutes phylum, Clostridia sp. was found both in the flowing (band 10) and the stationary phase (band 19) implying its important role in the removal of organic waste in the MRIS. Two bands (band 5 and 6) belonging to S. barnesii SES-3 were also found. S. barnesii is frequently found in wastewater to simultaneously remove selenite and nitrate [37], [38]. Only one band (15) directly relative to nitrification was found (Figure 4), which shared 97% sequence similarity to N. moscoviensis NSP M-1. However, no band sharing high similarity to ammonia oxidizers was found. This might be attributed to the population of AOB is too less to be detected, or the primers used for universal bacterial 16S rRNA genes did not match well with AOB template.
Due to no band retrieved from DGGE profile of bacterial 16S rRNA gene showed sequence similarity to ammonia oxidizers, making it difficult to elucidate the abundance and diversity of ammonia oxidizers. Therefore detection of the amoA gene encoding the subunit of AMO was necessary. BLAST analysis of AOB amoA genes found that band 1, band 2, band 5 shared 95, 99, and 96% sequence similarity to Nitrosospira, respectively (Figure 3B and Table S1). Nitrosospira sp. was vital for ammonia-oxidizing activity in acid soils, the community shift of Nitrosospira population was found to be pH-associated [32], [33]. Though the pH value of the stationary phase (P1–8) had not yet been detected in this article, the niche of P5–8 might be acidic because pH in the flowing phase dramatically decreased along the depth of the filter sheet (Figure 2B). Actually, a pH-associated trend might exist as implied by the distinctive appearance of these bands: band 1 was found only in P5 and P6; band 2 was found in all samples, especially in P4 and P5; band 5 was found in P6 and P7.
From the AOM abundance we could draw the conclusion that both AOB and AOA contributed to nitrification in the MRIS. The presence of distinctive phylotypes and the relatively higher ratio of archaeal vs. bacterial transcriptional activity implied that autotrophic ammonia oxidation in this system might be attributed mainly to archaea. The indistinctive dominance of AOA over AOB might correlate with different microenvironments in different layers of the MRIS. Further research should be implemented to link various environment factors that shape the diversity and abundance of AOB and AOA.
Supporting Information
AOB amo A genes retrieved from DGGE profile.
(DOC)
AOA amo A genes retrieved from DGGE profile.
(DOC)
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. The sequences reported in this study have been deposited in GenBank under accession numbers JQ963286 to JQ963324 for 16S rRNA V3 region of bacterial and KF460097 to KF460108 for amoA genes of AOB and AOA.
Funding Statement
This study was partly funded by innovative Program of The Chinese Academy of Sciences (KZCX2-YW-JC105), the Chinese National Programs for High Technology Research and Development (863 Program) (2011AA060904). The assistance from the Teamwork Project of the Natural Science Foundation of Guangdong Province, China (9351007002000001) is greatly appreciated. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Schmidt I, Sliekers O, Schmid M, Bock E, Fuerst J, et al. (2003) New concepts of microbial treatment processes for the nitrogen removal in wastewater. FEMS Microbiol Rev 27:481–492. [DOI] [PubMed] [Google Scholar]
- 2. van der Wielen PW, Voost S, van der Kooij D (2009) Ammonia-oxidizing bacteria and archaea in groundwater treatment and drinking water distribution systems. Appl Environ Microbiol 75:4687–4695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. He JZ, Shen JP, Zhang LM, Zhu YG, Zheng YM, et al. (2007) Quantitative analyses of the abundance and composition of ammonia-oxidizing bacteria and ammonia-oxidizing archaea of a Chinese upland red soil under long-term fertilization practices. Environ Microbiol 9:2364–2374. [DOI] [PubMed] [Google Scholar]
- 4. Nicol GW, Schleper C (2006) Ammonia-oxidising Crenarchaeota: important players in the nitrogen cycle? Trends Microbiol 14:207–212. [DOI] [PubMed] [Google Scholar]
- 5. Treusch AH, Leininger S, Kletzin A, Schuster SC, Klenk HP, et al. (2005) Novel genes for nitrite reductase and Amo-related proteins indicate a role of uncultivated mesophilic crenarchaeota in nitrogen cycling. Environ Microbiol 7:1985–1995. [DOI] [PubMed] [Google Scholar]
- 6. Leininger S, Urich T, Schloter M, Schwark L, Qi J, et al. (2006) Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature 442:806–809. [DOI] [PubMed] [Google Scholar]
- 7. Offre P, Prosser JI, Nicol GW (2009) Growth of ammonia-oxidizing archaea in soil microcosms is inhibited by acetylene. FEMS Microbiol Ecol 70:99–108. [DOI] [PubMed] [Google Scholar]
- 8. Kim D-J, Lee D-I, Keller J (2006) Effect of temperature and free ammonia on nitrification and nitrite accumulation in landfill leachate and analysis of its nitrifying bacterial community by FISH. Bioresour Technol 97:459–468. [DOI] [PubMed] [Google Scholar]
- 9. Jones R, Hood MA (1980) Effects of temperature, pH, salinity, and inorganic nitrogen on the rate of ammonium oxidation by nitrifiers isolated from wetland environments. Microbial Ecol 6:339–347. [DOI] [PubMed] [Google Scholar]
- 10. Erguder TH, Boon N, Wittebolle L, Marzorati M, Verstraete W (2009) Environmental factors shaping the ecological niches of ammonia-oxidizing archaea. FEMS Microbiol Rev 33:855–869. [DOI] [PubMed] [Google Scholar]
- 11. Purkhold U, Pommerening-Röser A, Juretschko S, Schmid MC, Koops H-P, et al. (2000) Phylogeny of all recognized species of ammonia oxidizers based on comparative 16S rRNA and amoA sequence analysis: implications for molecular diversity surveys. Appl Environ Microbiol 66:5368–5382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Boon N, Windt W, Verstraete W, Top EM (2002) Evaluation of nested PCR-DGGE (denaturing gradient gel electrophoresis) with group-specific 16S rRNA primers for the analysis of bacterial communities from different wastewater treatment plants. FEMS Microbiol Ecol 39:101–112. [DOI] [PubMed] [Google Scholar]
- 13.Apha A (1998) Standard methods for the examination of water and wastewater. Washington, DC: American Public Health Association; American Water Works Association; and Water Pollution Control Federation.
- 14. Brosius J, Dull TJ, Noller HF (1980) Complete nucleotide sequence of a 23S ribosomal RNA gene from Escherichia coli . Pro Natl Acad Sci 77:201–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ovreås L, Forney L, Daae FL, Torsvik V (1997) Distribution of bacterioplankton in meromictic Lake Saelenvannet, as determined by denaturing gradient gel electrophoresis of PCR-amplified gene fragments coding for 16S rRNA. Appl Environ Microbiol 63:3367–3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rotthauwe J-H, Witzel K-P, Liesack W (1997) The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl Environ Microbiol 63:4704–4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tourna M, Freitag TE, Nicol GW, Prosser JI (2008) Growth, activity and temperature responses of ammonia-oxidizing archaea and bacteria in soil microcosms. Environ Microbiol 10:1357–1364. [DOI] [PubMed] [Google Scholar]
- 18. Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599. [DOI] [PubMed] [Google Scholar]
- 19. Jia Z, Conrad R (2009) Bacteria rather than Archaea dominate microbial ammonia oxidation in an agricultural soil. Environ Microbiol 11:1658–1671. [DOI] [PubMed] [Google Scholar]
- 20. Mincer TJ, Church MJ, Taylor LT, Preston C, Karl DM, et al. (2007) Quantitative distribution of presumptive archaeal and bacterial nitrifiers in Monterey Bay and the North Pacific Subtropical Gyre. Environ Microbiol 9:1162–1175. [DOI] [PubMed] [Google Scholar]
- 21.Auset M, Keller AA, Brissaud F, Lazarova V (2005) Intermittent filtration of bacteria and colloids in porous media. Water Resour Res 41.
- 22. Chain P, Lamerdin J, Larimer F, Regala W, Lao V, et al. (2003) Complete genome sequence of the ammonia-oxidizing bacterium and obligate chemolithoautotroph Nitrosomonas europaea . J Bacteriol 185:2759–2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wendroth O (2004) Multivariate analysis of ecological data using CANOCO. Vadose Zone J 3:1057–1057. [Google Scholar]
- 24.Koops H-P, Purkhold U, Pommerening-Röser A, Timmermann G, Wagner M (2006) The lithoautotrophic ammonia-oxidizing bacteria. The prokaryotes: Springer. 778–811. [Google Scholar]
- 25. Ruiz G, Jeison D, Charmy R (2003) Nitrification with high nitrite accumulation for the treatment of wastewater with high ammonia concentration. Water Res 37:1371–1377. [DOI] [PubMed] [Google Scholar]
- 26. Gieseke A, Tarre S, Green M, De Beer D (2006) Nitrification in a biofilm at low pH values: role of in situ microenvironments and acid tolerance. Appl Environ Microbiol 72:4283–4292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Boyle-Yarwood SA, Bottomley PJ, Myrold DD (2008) Community composition of ammonia-oxidizing bacteria and archaea in soils under stands of red alder and Douglas fir in Oregon. Environ Microbiol 10:2956–2965. [DOI] [PubMed] [Google Scholar]
- 28. Yoo H, Ahn K-H, Lee H-J, Lee K-H, Kwak Y-J, et al. (1999) Nitrogen removal from synthetic wastewater by simultaneous nitrification and denitrification (SND) via nitrite in an intermittently-aerated reactor. Water Res 33:145–154. [Google Scholar]
- 29. Mota C, Head MA, Ridenoure JA, Cheng JJ, Francis L (2005) Effects of aeration cycles on nitrifying bacterial populations and nitrogen removal in intermittently aerated reactors. Appl Environ Microbiol 71:8565–8572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Koops H-P, Pommerening-Röser A (2001) Distribution and ecophysiology of the nitrifying bacteria emphasizing cultured species. FEMS Microbiol Ecol 37:1–9. [Google Scholar]
- 31. Hatzenpichler R, Lebedeva EV, Spieck E, Stoecker K, Richter A, et al. (2008) A moderately thermophilic ammonia-oxidizing crenarchaeote from a hot spring. Pro Natl Acad Sci 105:2134–2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Di HJ, Cameron KC, Shen J-P, Winefield CS, O'Callaghan M, et al. (2010) Ammonia-oxidizing bacteria and archaea grow under contrasting soil nitrogen conditions. FEMS Microbiol Ecol 72:386–394. [DOI] [PubMed] [Google Scholar]
- 33. Francis CA, Roberts KJ, Beman JM, Santoro AE, Oakley BB (2005) Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. Pro Natl Acad Sci 102:14683–14688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bock E, Schmidt I, Stüven R, Zart D (1995) Nitrogen loss caused by denitrifying Nitrosomonas cells using ammonium or hydrogen as electron donors and nitrite as electron acceptor. Arch Microbiol 163:16–20. [Google Scholar]
- 35. Könneke M, Bernhard AE, José R, Walker CB, Waterbury JB, et al. (2005) Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 437:543–546. [DOI] [PubMed] [Google Scholar]
- 36. Park SJ, Yoon JC, Shin K, Kim EH, Yim S, et al. (2007) Dominance of endospore-forming bacteria on a Rotating Activated Bacillus Contactor biofilm for advanced wastewater treatment. J Microbiol 45:113. [PubMed] [Google Scholar]
- 37. Lenz M, Enright AM, O’Flaherty V, van Aelst AC, Lens PN (2009) Bioaugmentation of UASB reactors with immobilized Sulfurospirillum barnesii for simultaneous selenate and nitrate removal. Appl Microbiol Biot 83:377–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mohanakrishnan J, Kofoed MVW, Barr J, Yuan Z, Schramm A, et al. (2011) Dynamic microbial response of sulfidogenic wastewater biofilm to nitrate. Appl Microbiol Biot 91:1647–1657. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
AOB amo A genes retrieved from DGGE profile.
(DOC)
AOA amo A genes retrieved from DGGE profile.
(DOC)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. The sequences reported in this study have been deposited in GenBank under accession numbers JQ963286 to JQ963324 for 16S rRNA V3 region of bacterial and KF460097 to KF460108 for amoA genes of AOB and AOA.