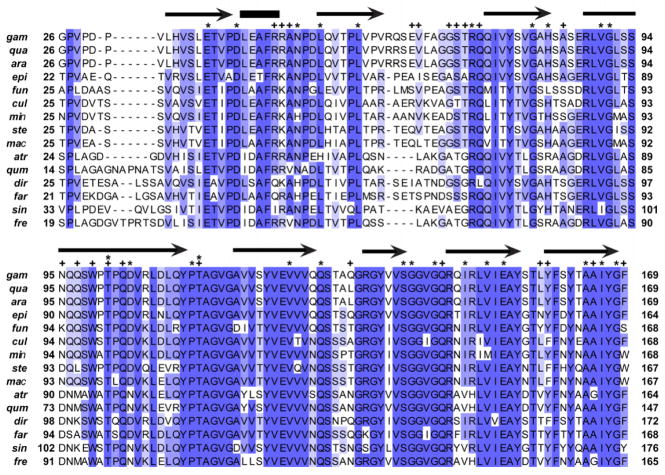
Figure 1. Amino acid alignment of AgSGU with putative orthologs from a sample of anopheline mosquitoes.
Shading corresponds to different levels of amino acid identity: dark, 13–15 out of 15 conserved (> 80% identity); intermediate, 10–12 out of 15 conserved (66.7%–80% identity); light = 7–9 out of 15 conserved (46.7%–66.6% identity). Above the alignment, arrows and bars represent annotations based on predictions of secondary structure, where arrows are predicted beta strands and bars are predicted alpha helices. Stars represent sites under negative selection identified by two separate methods (SLAC and FEL), while pluses indicate amino acids predicted to take part in protein binding. All species are in the genus Anopheles with abbreviations of the species names as follows: gam, gambiae; qua, quadriannulatus A; ara, arabiensis; epi, epiroticus; fun, funestus; cul, culicifacies A; min, minimus A; ste, stephensi; mac, maculatus B; atr, atroparvus; qum, quadrimaculatus; dir, dirus A; far, farauti; sin, sinensis; fre, freeborni.