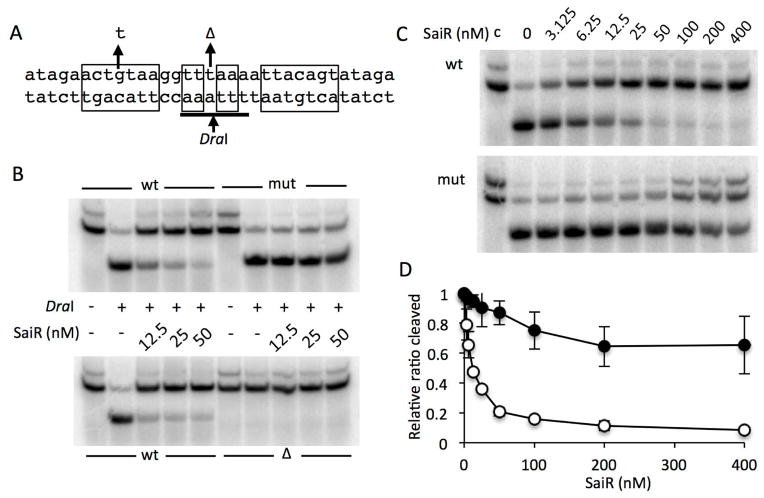
Fig. 5. SaiR directly interacts with the spxA2 core promoter.
A. The probes used for DraI-digestion protection assay. The sequence from −28 to +5 (relative to the transcriptional start site) of the spxA2 promoter is shown. The nucleotide sequences in rectangular boxes indicate the dyad symmetry required for SaiR repression. The arrows above the sequence show a base substitution and deletion used in the protection assay. The DraI recognition sequence is underlined and digestion site is marked by the arrow below the sequence.
B. The wild-type probe and the probe carrying the base substitution (mut) and deletion (Δ) were incubated without or with various concentrations of SaiR-His6, followed by DraI digestion. The results are representative of four independent experiments.
C. Titration of SaiR for protection against DraI digestion. All lanes except those marked with c were from reactions treated with DraI as described in Experimental procedures.
D. Results in triplicate of reactions shown in Fig. 5C were used to quantify band intensities using Image J. First, intensity of digested band in each lane was divided by the sum of digested and undigested band intensities in the corresponding lane (ratio cleaved). The relative ratio cleaved was calculated by dividing the ratio cleaved in each lane by the ratio obtained from the lane without SaiR. Symbols: open circles, the wild-type probe; closed circles, the probe with the base substitution.