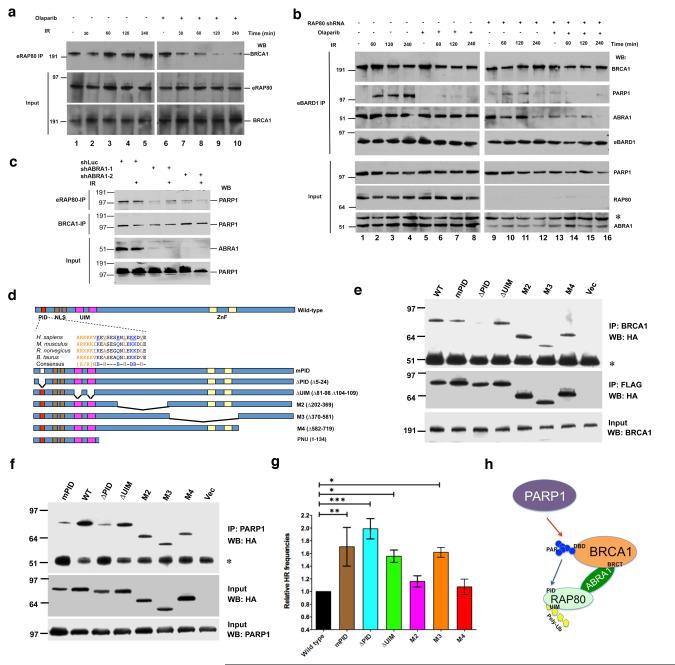
Figure 3. The interaction between PARsylated BRCA1 and RAP80 is required for maintaining BRCA1-RAP80-PARP1 complex integrity after DNA damage and normal HRR regulation.
(a) HeLa cells stably expressing FLAG/HA-tagged RAP80 (eRAP80) were incubated with 30 nM Olaparib for 48 hours and then irradiated with 10 Gy IR. Cells were lysed 30 min, 60 min, 120 min or 240 min post-IR. IPs were performed with antibodies directed against FLAG (recognizing eRAP80) and then probed with anti-BRCA1. Input samples were blotted and probed with an anti-HA (recognizing eRAP80) or an anti-BRCA1 antibody. (b) HeLa cells that stably express FLAG/HA-tagged BARD1 (eBARD1) and an shRNA containing either a RAP80-targeting or a control sequence were generated. Where indicated, these cells were incubated with 30 nM Olaparib or DMSO for 48 hours. Cells were then irradiated with 10 Gy IR and lysed 60 min, 120 min or 240 min later. IPs were performed with anti-FLAG antibody (recognizing eBARD1) and then probed with antibodies directed against BRCA1, PARP1 or ABRA1. Input proteins were blotted and probed with antibodies against PARP1, RAP80 or ABRA1. The asterisk in the ABRA1 blot indicates the migration position of a non-specific band detectable by this antibody. (c) HeLa cells stably expressing eRAP80 were infected with a lentivirus encoding an shRNA directed at Luciferase (shLuc) or ABRA1 (shABRA-1 or shABRA-2). 48 hours later, cells were irradiated with 10 Gy IR or mock treated. Cells were collected 4 hours after IR. IPs were performed with antibodies directed at FLAG (recognizing eRAP80) or BRCA1 and then probed with anti-PARP1 antibody. Input samples were blotted and probed with an anti-ABRA1 or an anti-PARP1 antibody. (d) RAP80 contains a PAR interacting domain. Schematic diagrams indicate the domain structure of human RAP80. PID, poly-ADP-ribose interacting domain; NLS, nuclear localization sequence; UIM, ubiquitin interacting motif; and ZnF, Zinc finger domain. The sequences of the PID domain in four mammalian species are shown. The conserved K/R-rich cluster, hydrophobic, and basic amino acids are indicated in different colors. Seven conserved amino acids (underlined) were mutated to alanine [A] in the mPID mutant. Structures of six N-terminal GST or C-terminal FLAG/HA-tagged RAP80 mutants and an N-terminal T7-tagged RAP80 PNU fragment are shown below. (e) 293T cells were transiently transfected with the RAP80-FLAG/HA vectors described in (d) and, 72 hours later, cells were irradiated with 10 Gy IR. Cells were lysed 3 hours later, and IPs were generated with an anti- BRCA1 antibody, and the IPd proteins were blotted and the blots probed with an anti-HA (top panel and middle panels) antibody. The asterisk indicates the position of IgG heavy chain. 15 μg of protein from each lysate were loaded onto each gel lane, and the subsequent blots were probed with an anti-BRCA1 antibody (bottom panel). (f) 293T cells were treated as in (e). IPs were performed with an anti-PARP1 antibody. IPd proteins were blotted and probed with an anti-HA antibody to recognize epitope-tagged RAP80 mutants (top panel). The asterisk indicates the migration position of IgG heavy chain. 15 μg of protein from each lysate were loaded onto each gel lane, and the subsequent blots were probed with anti-HA (middle panel) or anti-PARP1 antibody (bottom panel). (g) A graph summarizing relative HRR frequencies of cells over-expressing wild-type RAP80 or various RAP80 mutants. DR-GFP HRR assays were performed as described in methods. The abundance of GFP-positive (HRR) cells over-expressing wild-type RAP80 was normalized to one. Experiments were performed in triplicate, and error bars indicate standard deviation. *, p<0.05; **, p<0.01; ***, p<0.001. (h) A schematic diagram showing the relationship between PARP1, BRCA1, ABRA1 and RAP80 in the HRR-tuning complex. Red arrow indicates that PARP1 promotes BRCA1 PARsylation. The blue arrow indicates the binding of PAR to the RAP80 PID domain. BRCA1 BRCT and DNA-binding (DBD) domains, and RAP80 UIM domain are also indicated in the diagram.