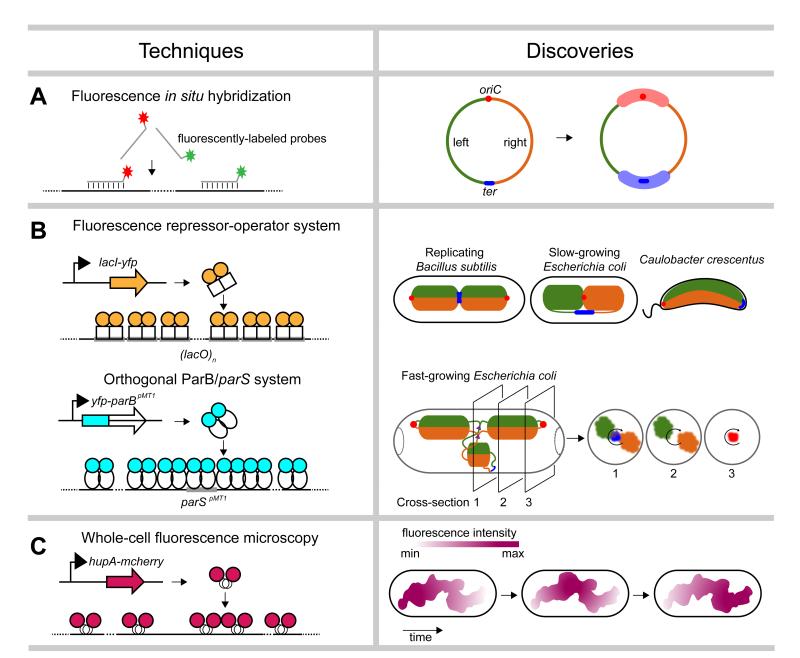
Figure 1. Single-cell microscopic techniques.
A summary of microscopic techniques (left) used to probe the spatial organization of bacterial chromosomes, with example discoveries (right). (a) Fluorescence in situ hybridization. The origin of replication (oriC) is shown as a red dot and the terminus (ter) as a blue line. The left and right arms of the chromosome are colored in green and orange, respectively. The Ori and Ter macrodomains are indicated by thick opaque red and blue lines, respectively. (b) Fluorescent repressor-operator systems. Proposed patterns of chromosome organization in replicating Bacillus subtilis, slow-growing Escherichia coli, and Caulobacter crescentus based on traditional fluorescent repressor-operator systems (top). The proposed spatial organization of replicating chromosomes in fast-growing E. coli cells, based on studies using an orthogonal ParB/parS system is also shown (bottom). Cross-sections show the radial distribution of the left and right chromosomal arms as well as oriC, ter, and replication forks (purple triangles). (c) Imaging of fluorescently-tagged nucleoid-associated proteins. Reconstruction of the nucleoid of slow-growing E. coli cells expressing HupA-mCherry indicated a coiled-shaped structure of no particular handedness. Nucleoid intensities, summarized from Z projections, also showed rapid fluxes of density along the length of the nucleoid.