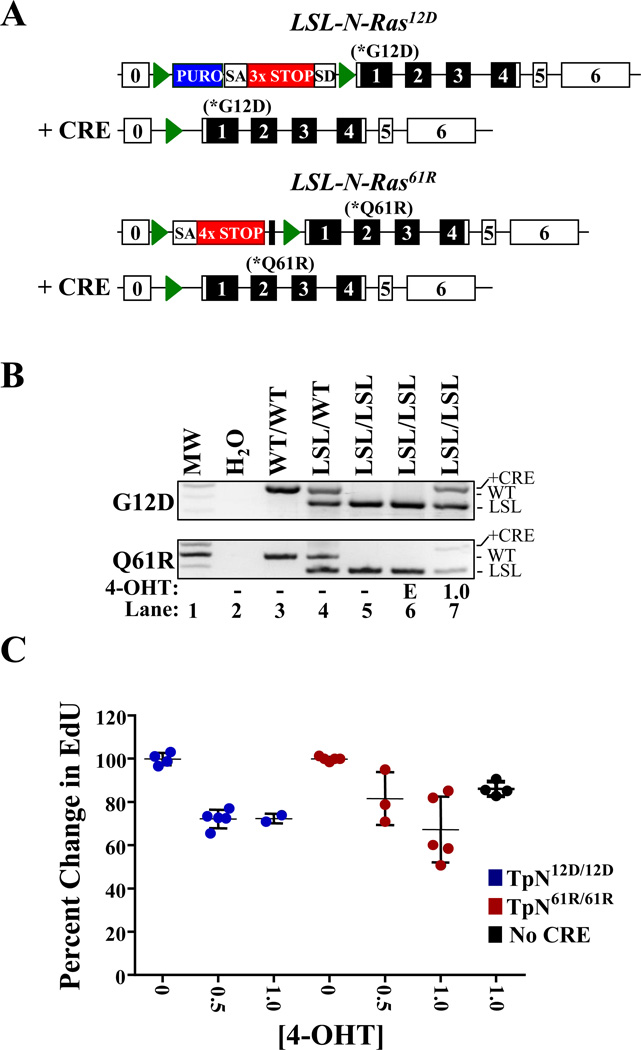
Figure 1. Activation of LSL-N-RasQ61R or LSL-N-RASG12D slows melanocyte proliferation.
(A) Diagrammatic representation of the LSL-N-RASG12D (17) and LSL-N-RasQ61R alleles in the presence or absence of CRE recombinase. Green triangles denote lox P sites and unnumbered black boxes represent FRT sites. Blue boxes indicate the location of a puromycin (PURO) resistance cassette. SA-splice acceptor; SD-splice donor; 3×STOP- transcription and translational stop sequence. (B) PCR genotyping of melanocytes treated for 6 days with ethanol vehicle (E; lane 7) or 1.0 µM 4-OHT (lane 7). DNA from mice of the indicated genotypes is included as a control (lanes 3–5). (C) Melanocytes cultured as in ‘B’ were incubated with EdU, a thymidine analog, for 16 hours and then analyzed by flow cytometry. EdU incorporation in vehicle treated cells was set to 100%. Each dot represents a biological replicate with the mean and standard error of the mean indicated by black lines.