Fig. 1.
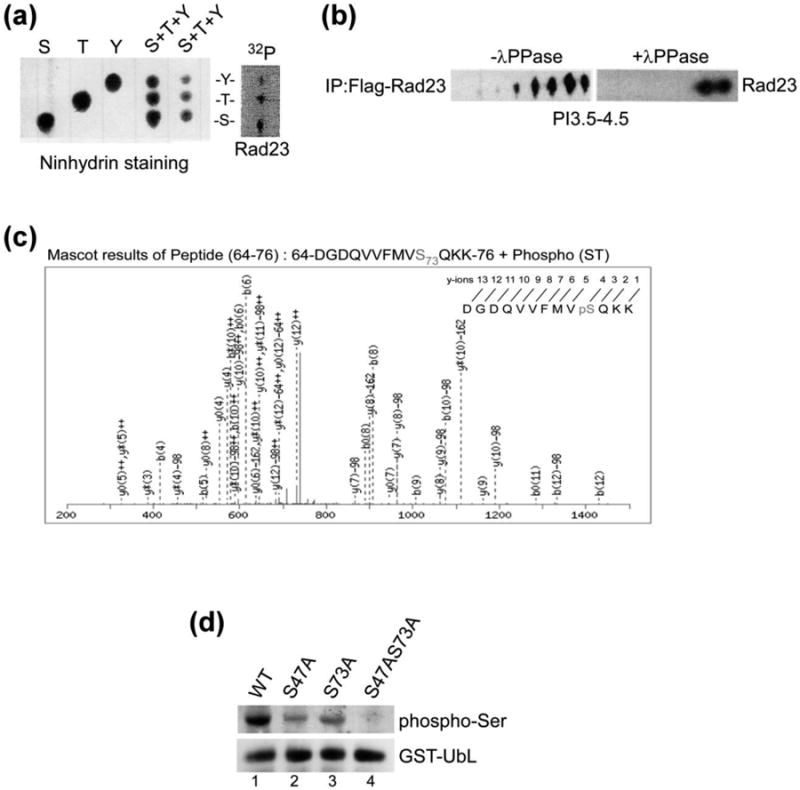
Rad23 are phosphorylated at multiple residues in vivo. (a) Yeast cells expressing Flag-Rad23 was affinity purified from exponential-phase yeast cells that were incubated with [32P]-orthophosphate for 1 h. The in vivo phosphorylated Flag-Rad23 was separated by SDS-PAGE, and transferred to PVDF membrane. 32P-labeled Flag-Rad23 was identified by autoradiography, excised and digested in acid. The hydrolysate was separated by one-dimensional thin-layer chromatography (TLC). The positions of non-radioactive phosphoamino acid standards were detected by staining with ninhydrin (left panel). The independent standards and two different amounts of a mixture of the three combined standards are shown. The pattern of radioactive spots generated from the hydrolyzed of 32P-Flag-Rad23 is shown on the right (Flag-Rad23), and the positions of phosphorylated tyrosine (Y), threonine (T), and serine (S) residues are indicated. (b) Flag-Rad23 was immunoprecipitated from yeast cells, and incubated with or without λ-phosphatase for 1 h. Proteins were released from the affinity beads and resolved by isoelectric focusing (IEF). The separated proteins were resolved in the second dimension in SDS-PAGE, transferred to nitrocellulose and incubated with anti-Flag antibody. (c) Recombinant GST-Rad23 was purified from E. coli BL21 (DE3) cells, extensively washed by lysis buffer and subjected to in vitro kinase assay followed by LC-MS/MS analysis. (d) GST-UbL and mutants with changes in candidate phosphorylation sites were isolated from rad23Δ strain and in vivo phosphorylation was investigated by immunoblotting, using antibodies that recognize phospho-serine residues.
