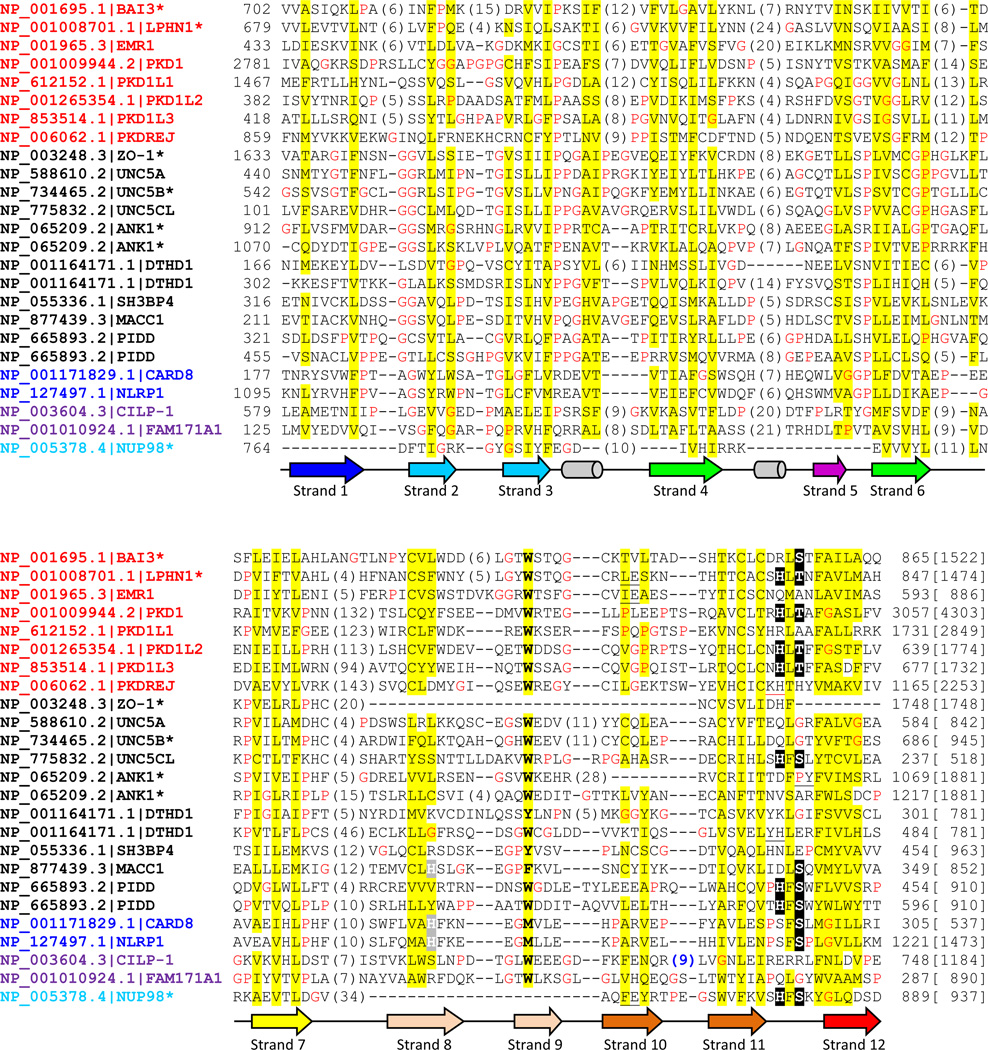
Figure. 2. Multiple sequence alignment of selected human proteins containing GAIN, ZU5 and Nup98 C-terminal domains.
Human proteins found in transitive PSI-BLAST searches were selected including several GPCR sequences. Multiple sequence alignment was built by PROMALS3D [55] and edited manually. Common gene names following NCBI Genbank accession numbers are used for each protein. Names of GAIN, canonical ZU5, ZU5-FIIND, ZU5-CF and Nup98 C-terminal domain are in red, black, blue, magenta and cyan colors, respectively. An asterisk is labeled where the protein or one of its close homologs has available structures. The conserved serine, threonine and histidine in the cleavage site are highlighted in black background and putative active site histidines in beta-strand 8 in ZU5-FIIND and MACC1 are shaded in grey. The protein length is noted in brackets at the end of the alignment. Nonpolar residues in mainly hydrophobic positions are highlighted in yellow. Glycines and prolines are colored in red. The column of mainly aromatic residue is in bold. Secondary structures are represented as arrows (beta-strands) and tubes (alpha-helices) below and colored consistently with structures in Fig. 1. Insertions are represented by the numbers of inserted residues in parentheses. Several one residue insertions are black underscored and the red underscore in PKDREJ represents a 19-residue insertion before the deteriorated cleavage motif. And the insertion in CILP-1 (sequence: RNKREDRTF) with a consensus furin-like cleavage site is colored in blue.