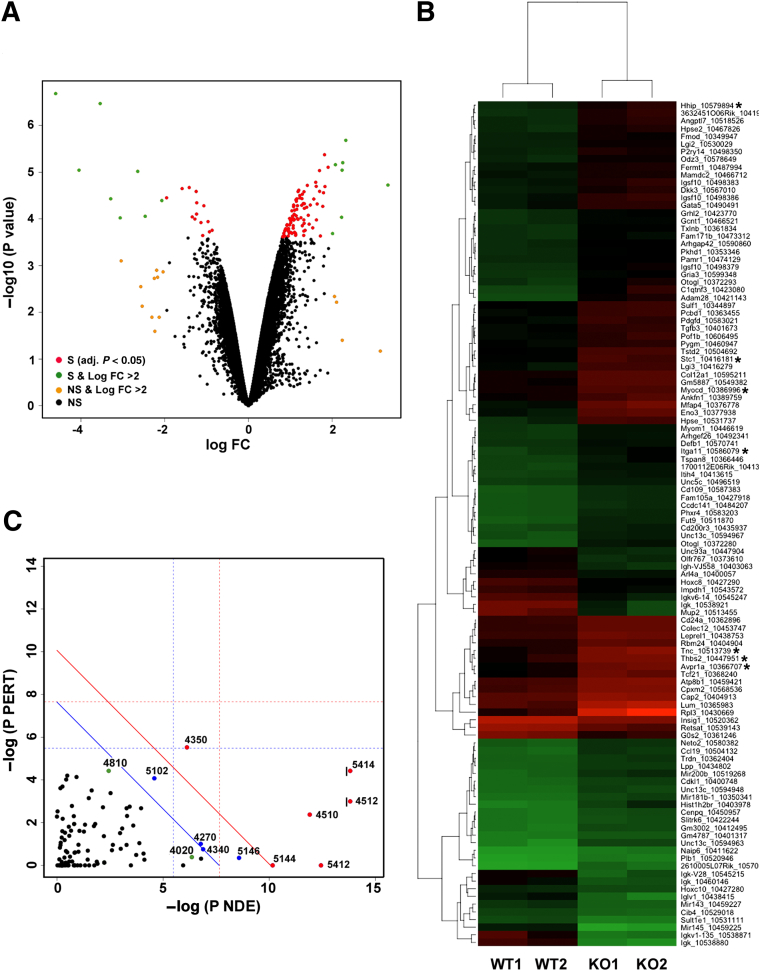
Figure 5.
Gene expression profiling in ureters from WT and miR-143/145 KO mice. Total RNA samples from WT and mutant ureters were analyzed using Affymetrix GeneChip Mouse Gene 1.0 ST arrays. A: Volcano plot shows the fold-difference in the expression and the significance of the difference for all genes in the arrays. B: Heatmap shows differentially expressed genes in ureters from miR-143/145 KO mice. Of the 108 differentially expressed genes, 90 were up-regulated and 18 were down-regulated. Each column represents results from an independent animal (indicated as WT1, WT2, KO1, and KO2). Each row corresponds to a single gene probe. The color indicates the level of mRNA expression (red, high expression; green, low expression). We used hierarchical cluster analysis of gene expression to group genes with similar expression patterns as indicated at the left side of the map. C: SPIA of genes showing differential expression in miR-143/145 KO ureters. A two-dimensional plot shows the relationship between the gene over-representation (x axis) and the perturbation of the pathways (y axis) analyzed by SPIA. We considered the 500 most differentially expressed genes for the analysis. Each point represents a pathway. The x axis shows the P value on the over-representation of differentially expressed genes in the given pathway: P(NDE). The y axis shows the significance of the overall perturbation of the pathway: P(PERT). Pathways above the red line are significant at 5% after Bonferroni correction, and pathways above the blue line are significant at 5% after FDR correction. The vertical and horizontal thresholds represent the same corrections for the two types of evidence considered individually. Indicated in green are relevant pathways that did not meet both the over-representation and perturbation criteria but were above a 10% cut-off of the FDR-corrected overall P value: P(G)FDR. For pathways 4512 and 5414, the actual −log P(NDE) values were 22 and 14.5, respectively. These values were cut off from the plot as indicated by the black vertical line on the left side of the point. Numbers are the Kyoto Encyclopedia of Genes and Genomes (Kegg) database identification number for the pathway: 4512, ECM-receptor interaction; 5414, dilated cardiomyopathy; 4510, focal adhesion; 5412, arrhythmogenic right ventricular cardiomyopathy; 4350, transforming growth factor-β signaling pathway; 4270, vascular smooth muscle contraction; 4340, hedgehog signaling pathway; 4810, regulation of actin cytoskeleton; 4020, calcium signaling pathway; 5146, amebiasis; 5144, malaria; and 5202, transcriptional dysregulation in cancer. Genes related to smooth muscle biology are marked with asterisks. FC, fold-change; NS, not significant (P > 0.05); S, significant (P < 0.05).