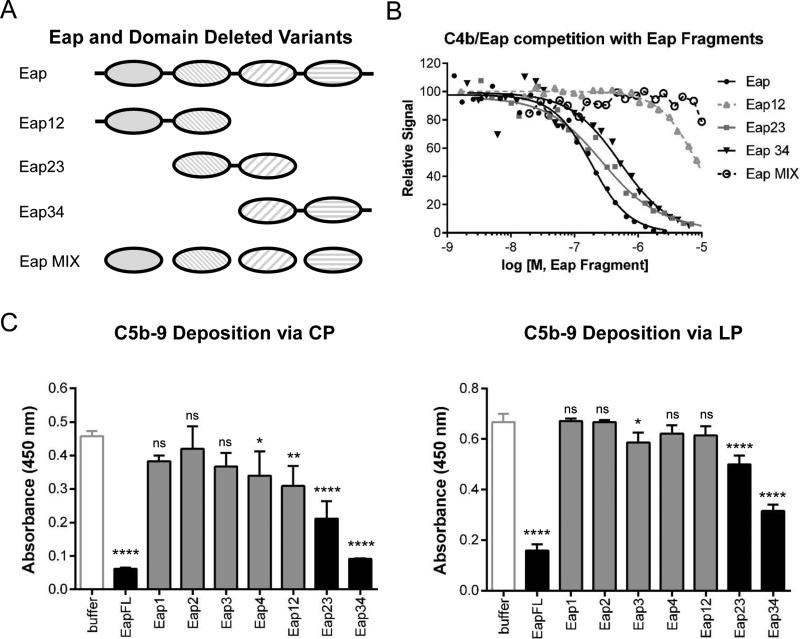
Figure 4. The Third Domain of Eap is Necessary for C4b Binding and Inhibition of the CP/LP.
(a) Diagram of full-length and domain-deleted forms of Eap used to map functional sites within the Eap protein. (b) The ability of untagged Eap, Eap12, Eap23, Eap34, and an equimolar mixture of individual Eap domains (Eap MIX) to compete the AlphaScreen signal generated by myc-Eap and C4b-biotin was assessed over a logarithmic dilution series. While three independent trials were carried out, the data presented here are from a single representative titrations. The smooth line indicates the outcome of fitting all points to a dose-response curve when competition was observed. Legend is inset. (c) The effect of including 1 μM Eap, or various truncations thereof, on C5b-9 deposition on ELISA plates coated with either CP- (left panel) or LP-specific (right panel) activators. 1% (v/v) NHS was used as a source of complement components. Error bars represent the mean ± standard deviation of three independent experiments. Measures of statistical significance were determined by one-way ANOVA for the various Eap truncations versus buffer control alone. *, p≤0.05; **, p≤0.01; ****, p≤0.0001; ns, not significant.