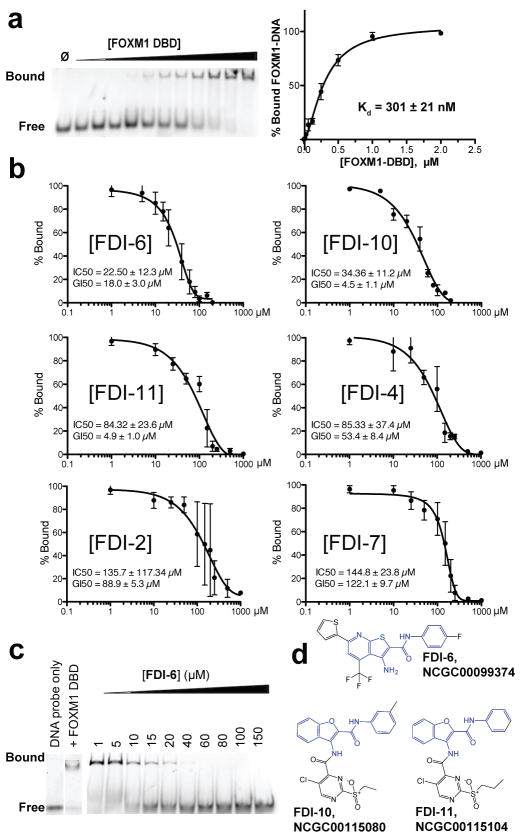
Figure 2. EMSA provides orthogonal biophysical validation of 16 lead compounds from FP screen.
(a) Representative EMSA image shows the association of FOXM1 DBD with its fluorescently tagged DNA consensus. Binding curve for the FOXM1 DBD-DNA interaction was determined by quantification of EMSA replicates. (b) Quantification of EMSA bands confirmed six of the 20 lead compounds selected from the FP results inhibited FOXM1 DNA binding. In rank order of potencies: FDI-6 >FDI-10 > FDI-11 > FDI-4 > FDI-2 > FDI-7. Representative gel images are provided in Supplementary concentrations in MCF-7 cells. (c) Representative Fig. 5. Each inset lists the IC50 as well as 72 h GI50 EMSA showing the inhibitory effect of FDI-6 on the FOXM1 DBD-DNA complex. (d) Structures of FDI-6, FDI-10, and FDI-11, the most potent of the validated hit compounds, which all contain a common core structural element (highlighted in blue). Data are reported as average of replicates (n=3) and error bars indicate s.d. throughout figure.