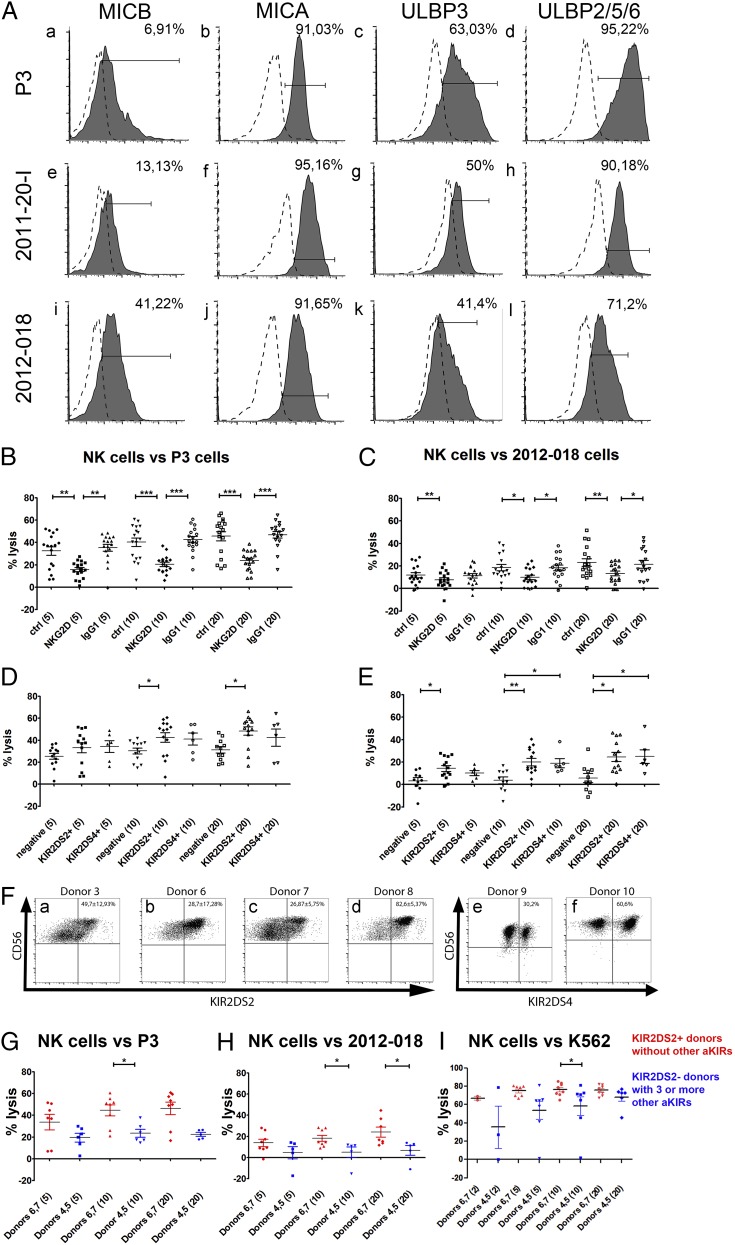
FIGURE 1.
Ligation of NKG2D to stress-induced ligands and the activating KIR2DS2 receptor are important for efficient NK cell–mediated cytotoxicity of GBM cells. (A) Surface expression of ligands for NKG2D receptors MICB, MICA, ULBP3, and ULBP 2/5/6 on P3 (Aa–Ad), 2011-20-I (Ae–Ah), and 2012-018 (Ai–Al) cells. Dashed graphs represent negative controls; filled graphs represent staining. The proportion of specifically stained cells indicated was calculated by subtracting the percentage of isotype-control–positive cells from the percentage of marker-positive cells. The percentage lysis by NK cells of P3 (B) and 2012-018 (C) GBM cells at E:T ratios of 5:1, 10:1, and 20:1, with and without addition of blocking NKG2D or IgG1 isotype-control Abs. Mean percentage (± SEM) of lysis of P3 (D) and 2012-018 (E) GBM cells mediated by KIR2DS2+ or KIR2DS4+ NK cells compared with NK cells lacking both receptors (negative) at E:T ratios of 5:1, 10:1, and 20:1. Data are mean ± SEM of three or four independent experiments/donor (n = 6 donors). The x-axis shows experimental condition and E:T ratio (in parentheses). *p < 0.05, **p = 0.01, ***p < 0.001. (F) CD56 versus KIR2DS2 dot plots represent the gating strategy of KIR2DS2+ subpopulations of donors 3 (Fa), 6 (Fb), 7 (Fc), and 8 (Fd). CD56 versus KIR2DS4 dot plots represent the gating strategy for KIR2DS4+ expression on NK cell subpopulations of donors 9 (Fe) and 10 (Ff). Data are mean percentage (± SEM) of cells expressing KIR2DS2 or KIR2DS4 receptors. Mean percentage (± SEM) of lysis of P3 GBM cells (G), 2012-018 GBM cells (H), and K562 cells (I) mediated by NK cells from donors 6 and 7 (red: possessing KIR2DS2 gene and no other activating KIR genes) compared with NK cells from donors 4 and 5 (blue: lacking KIR2DS2 gene, but possessing three or more genes of other activating KIRs) at E:T ratios of 2:1, 5:1, 10:1, and 20:1. Data are mean ± SEM of three or four independent experiments/donor. *p < 0.05.