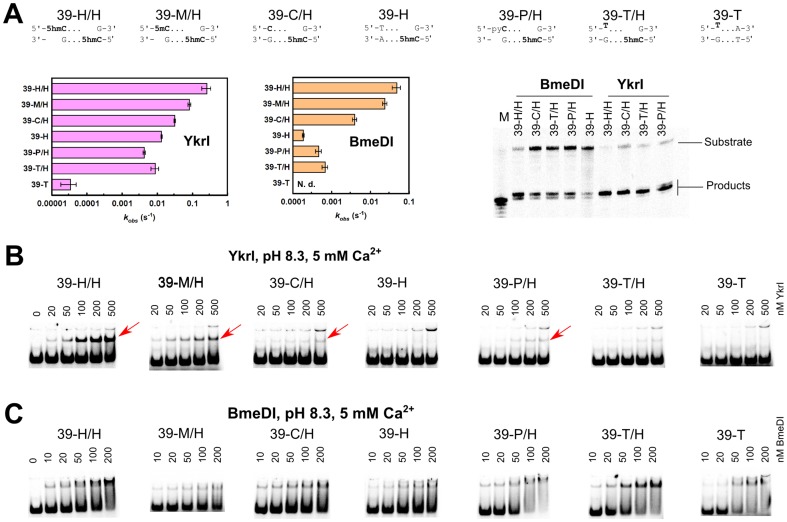
Figure 3. DNA cleavage and binding by YkrI and BmeDI.
The sequences at the top of the figure schematically depict the 39-H/H (optimal substrate with two 5hmC bases), 39-M/H, 39-C/H, 39-H, 39-P/H, 39-T/H, and 39-T (one or both 5hmC-G base pairs replaced with a 5mC-G, C-G, T-A, pyrrolocytosine-G, and thymine-G base pairs, Table 1) oligoduplexes. (A) The observed first-order DNA cleavage rate constants. Cleavage reactions were performed with 1 nM substrate and 100 nM enzyme (monomer) at 15°C. In our experimental setup, BmeDI cleavage of the 39-T oligoduplex was not detectable. Denaturing PAGE analysis of cleavage products formed with various DNA substrates is shown on the right-hand side. Gel lane ‘M’ contained a synthetic single-stranded oligonucleotide that corresponds to cleavage of the bottom strand 11 nt downstream of the 5hmC nucleotide. (B) Electrophoretic mobility shift assay with YkrI. DNA binding experiments were performed in a pH 8.3 buffer in the presence of 5 mM Ca2+ ions. The final DNA concentration was 1 nM, and YkrI concentrations are indicated above the gel lanes. Red arrows mark the location of the specific YkrI-DNA complexes. The upper band corresponds to the low-mobility non-specific YkrI-DNA complex formed due to binding/aggregation of multiple protein molecules. (C) Electrophoretic mobility shift experiments with BmeDI. Reaction conditions were as in panel (B).