Figure 1.
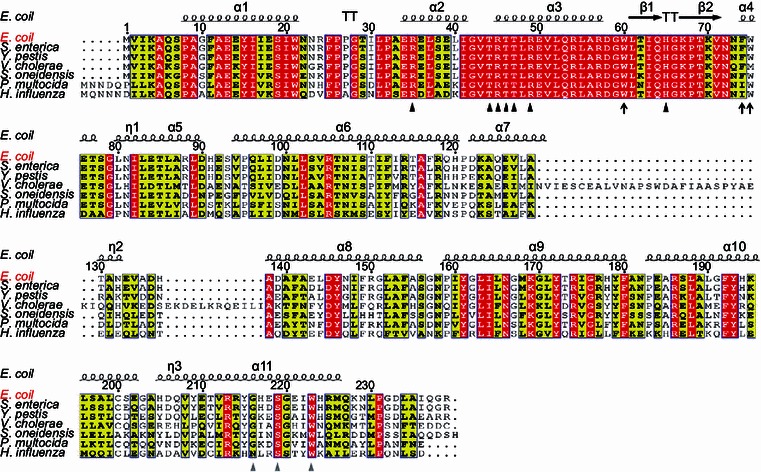
Sequence alignments ofE. coliFadR regulatory protein with other homologues from six different species of γ-proteobacteria. The multiple alignments of amino acid sequences were conducted using ClustalW2 (http://www.ebi.ac.uk/Tools/clustalw2/index.html), and the resultant output was processed by program ESPript 2.2 (http://espript.ibcp.fr/ESPript/cgi-bin/ESPript.cgi), generating the final BLAST photography (Feng & Cronan, 2011b). Identical residues are in white letters with red background, similar residues are in black letters in yellow background, and the varied residues are in grey letters. As we earlier described (Feng et al., 2008), the protein secondary structure was shown in cartoon (on top), in terms of the structural architecture of E. coli FadR protein (PDB:1E2X) (van Aalten et al., 2000). α: alpha-helix; β: beta-sheet; T: Turn; η: coil. The DNA binding sites are indicated with black triangles (R35, T44, R45, T46, T47, R49 and 65H) (Xu et al., 2001), the ligand binding sites are shown with grey triangles (216G, 219S and 223W) (van Aalten et al., 2001), and the newly-proposed amino acids essential for DNA binding activity of FadR protein are highlighted with dark arrows (W60, F74 and W75). The FadR sequences are separately sampled from E. coli K12 (Accession no.: CAA30881), S. enterica (Salmonella enterica) (Accession no.: ACF63827), and Y. pestis (Yersinia pestis) (Accession no.: ZP_06207826), V. cholerae (Vibrio cholerae) (Accession no.: AAO37924), S. oneidensis (Shewanella oneidensis) (Accession no.: NP_718457), P. multocida (Pasterurella multocida) (Accession no.: AAK02132), and H. influenza (Haemophilus influenza) (Accession no.: AAC22085)
